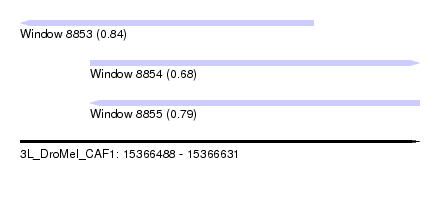
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,366,488 – 15,366,631 |
| Length | 143 |
| Max. P | 0.844337 |

| Location | 15,366,488 – 15,366,593 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
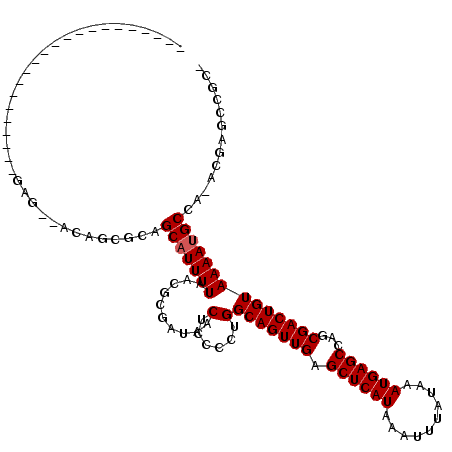
>3L_DroMel_CAF1 15366488 105 - 23771897 UUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCGACGACACUGAAACU---CGGU------------CGGAAAA ....((((............(((((((.((((((..........))))))...))))))).....((.......)))))).((((.(.......---.)))------------))..... ( -26.90) >DroVir_CAF1 96342 91 - 1 UUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCCA-ACGAGCCAU---------AUAACU---CGGU------------G----AA ....(((.............(((((((.((((((..........))))))...))))))).........-.((((....---------....))---))))------------)----.. ( -22.50) >DroPse_CAF1 99170 108 - 1 UUAAUGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCGACGACUCCG---CU---CGAG-----GG-GCCAUGAGCAG ....(((.......(((((.(((((((.((((((..........))))))...)))))))...........(((((.(.((....))))---))---))))-----))-)......))). ( -34.62) >DroMoj_CAF1 95017 91 - 1 UUAACACGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCCA-ACGAGCCGU---------AUAACU---CGCC------------G----AA ......((............(((((((.((((((..........))))))...))))))).........-.((((....---------....))---)).)------------)----.. ( -19.80) >DroAna_CAF1 233342 120 - 1 UUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCGACGACACUGAAAUUGGUCGGUUCGAGGGGGGCACGGAAAA ....((.(....(((((((.(((((((.((((((..........))))))...)))))))...........(((((((...(((........)))..))))))))))))))).))..... ( -39.50) >DroPer_CAF1 98690 108 - 1 UUAAUGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCGACGACUCCG---CU---CGAG-----GG-GCCAUGAGCAG ....(((.......(((((.(((((((.((((((..........))))))...)))))))...........(((((.(.((....))))---))---))))-----))-)......))). ( -34.62) >consensus UUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCGACGAC_C_G_AACU___CGGU____________CG___AA ....((((............(((((((.((((((..........))))))...))))))).....((.......))))))........................................ (-19.04 = -19.07 + 0.03)



| Location | 15,366,513 – 15,366,631 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15366513 118 + 23771897 CGCGGCUCGUUUCGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGCGUUAAAAUGCUUCGCUGU--UCCCACCGUAUUUUUCCUGGUUUUC .((((((.((..........))))))))((((((((..........)))))).))...((((((..((.....((((....))))..((.((.--...)).))...))..)))))).... ( -30.40) >DroVir_CAF1 96355 94 + 1 -AUGGCUCGU-UGGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGCGUUAAAAUGCUGCACGCU--CGC---------------------- -...((.(((-((((((((((....(((.(((((((..........))))))).((((.(....).))))...))).)))))))))).)))..--.))---------------------- ( -25.90) >DroGri_CAF1 97168 92 + 1 -ACGGCUCGU-UGGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGCGUUAAAAUGCUGC--GCU--CUG---------------------- -.(((..(((-.(((((((((....(((.(((((((..........))))))).((((.(....).))))...))).)))))))))))--)..--)))---------------------- ( -27.10) >DroMoj_CAF1 95030 94 + 1 -ACGGCUCGU-UGGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGUGUUAAAAUGCUGUGCUCU--CUG---------------------- -..(((..((-(((.......(((...)))((((((..........))))))))))).))).(((((((...(((......)))....)))))--)).---------------------- ( -25.40) >DroAna_CAF1 233382 118 + 1 CGCGGCUCGUUUCGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGCGUUAAAAUGCUUCGCUGUUUUCCCGAUGUACUUUCCUCCGC--CC .((((((.((..........))))))))((((((((..........)))))).))...((.(((..((((((((.(..(((..((...))..)))..)))))).)))..)))..))--.. ( -31.30) >DroPer_CAF1 98718 93 + 1 CGCGGCUCGUUUCGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGCAUUAAAAAGCUCCGCUGU--------------------------- .((((((.((..........))))))))((((((((..........)))))).)).....(((.((((((............)))))).))).--------------------------- ( -27.10) >consensus _ACGGCUCGU_UCGCAUUUUACAGUCGCUGGCUCAUUUAUAAAUUUAUGAGCUCAACUGCCAGGGGAGUUAUCGCGUUAAAAUGCUGCGCUCU__CCC______________________ .............((((((((....(((.(((((((..........))))))).((((.(....).))))...))).))))))))................................... (-21.06 = -21.25 + 0.19)



| Location | 15,366,513 – 15,366,631 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15366513 118 - 23771897 GAAAACCAGGAAAAAUACGGUGGGA--ACAGCGAAGCAUUUUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCG .....(((((.......((.((...--.)).))..((........)).........)))))((((((.((((((..........))))))...))))))......(((........))). ( -31.90) >DroVir_CAF1 96355 94 - 1 ----------------------GCG--AGCGUGCAGCAUUUUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCCA-ACGAGCCAU- ----------------------((.--..(((...((((((((.(((..(((((((...)).))))).((((((..........))))))..)))....))))))))..-))).))...- ( -26.30) >DroGri_CAF1 97168 92 - 1 ----------------------CAG--AGC--GCAGCAUUUUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCCA-ACGAGCCGU- ----------------------..(--..(--(..((((((((.(((..(((((((...)).))))).((((((..........))))))..)))....))))))))..-.))..)...- ( -22.30) >DroMoj_CAF1 95030 94 - 1 ----------------------CAG--AGAGCACAGCAUUUUAACACGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCCA-ACGAGCCGU- ----------------------...--.(.((.(.(((((((..((.(........).))(((((((.((((((..........))))))...))))))))))))))..-..).)))..- ( -20.60) >DroAna_CAF1 233382 118 - 1 GG--GCGGAGGAAAGUACAUCGGGAAAACAGCGAAGCAUUUUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCG ..--((((.(....(((((..((((.....(((...........)))......)))).))(((((((.((((((..........))))))...)))))))....)))....)...)))). ( -32.90) >DroPer_CAF1 98718 93 - 1 ---------------------------ACAGCGGAGCUUUUUAAUGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCGAAACGAGCCGCG ---------------------------.(((.((((................)))).)))(((((((.((((((..........))))))...))))))).....(((........))). ( -26.49) >consensus ______________________GAG__ACAGCGCAGCAUUUUAACGCGAUAACUCCCCUGGCAGUUGAGCUCAUAAAUUUAUAAAUGAGCCAGCGACUGUAAAAUGCCA_ACGAGCCGC_ ...................................(((((((..........(......)(((((((.((((((..........))))))...))))))))))))))............. (-19.95 = -20.12 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:24 2006