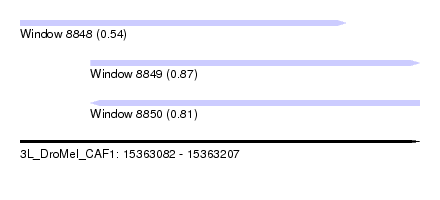
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,363,082 – 15,363,207 |
| Length | 125 |
| Max. P | 0.866529 |

| Location | 15,363,082 – 15,363,184 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.01 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -23.22 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.536004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15363082 102 + 23771897 ------------------CCUGCACAGCAUGCGAGUUUAUAGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAACGCGCUUUUUGCGGAUACGCUU ------------------.(((((.(((..(((.((((...((.(((....)))))....((((((.......))))))))))((((.....))))..))))))...)))))........ ( -24.70) >DroVir_CAF1 93113 102 + 1 ------------------UUUGCACAGCAUGCGGCUUUAUAGUCGAUUUUUAUCACAGCAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACGAUA ------------------((((((.(((..((((((((...((.(((....))))).....(((((.......))))))))))((((.....))))..))))))...))))))....... ( -25.80) >DroPse_CAF1 96482 102 + 1 ------------------UCCGCACAGCAUGCGAGUUUAUAGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACGCUU ------------------((((((.(((..((((((((...((.(((....)))))....((((((.......))))))))))((((.....)))).)))))))...))))))....... ( -29.40) >DroGri_CAF1 94118 102 + 1 ------------------UUUGCACAGCAUGCGGCUUUAUAGUCGAUUUUUAUCACAGCAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACAAUA ------------------((((((.(((..((((((((...((.(((....))))).....(((((.......))))))))))((((.....))))..))))))...))))))....... ( -25.80) >DroMoj_CAF1 91594 102 + 1 ------------------UUUGCACAGCAUGCGGCUUUAUAGUCGAUUUUUAUCACAGCGUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACGAUA ------------------((((((.(((...(((((....)))))............(((((((((.......))))))....((((.....))))..))))))...))))))....... ( -26.20) >DroAna_CAF1 230240 120 + 1 CUUUCAUCGAAAAAAAAACCCGCACAGCAUGCGAGUUUAUAGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAACGCGCUUUUUGCGGAUACGCUU .....(((((.....((((.((((.....)))).))))....)))))..........(((((((...........(((((((((((............)))))))))))))))))).... ( -29.30) >consensus __________________UCUGCACAGCAUGCGACUUUAUAGUCGAUUUUUAUCACAGCAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACGAUA ...................(((((.(((..(((........((.(((....))))).((.((((((.......))))))..))((((.....))))..))))))...)))))........ (-23.22 = -22.50 + -0.72)

| Location | 15,363,104 – 15,363,207 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -17.89 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15363104 103 + 23771897 AGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAACGCGCUUUUUGCGGAUACGCUUUAUU-------CUUUUU--UCCCCC---ACUUAUU----- .((.(((....)))))(((..(((((.......)))))(((((((((.....))))....(((.....))).....)))))...-------......--......---)))....----- ( -19.90) >DroPse_CAF1 96504 102 + 1 AGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACGCUUUAUU-------CUCCUU--CU-------ACUUAUUUCU-- .((.(((....)))))((((.(((((.......)))))(((((((((.....))))(((.(((.....))))))..)))))...-------......--.)-------))).......-- ( -22.90) >DroGri_CAF1 94140 108 + 1 AGUCGAUUUUUAUCACAGCAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACAAUAUUUU-------CAUUUUUUUUUUUCUAUUUUUGUC----- .((.(((....))))).((.((((((.......))))))..))((((.....))))(((.(((.....))))))..........-------........................----- ( -18.20) >DroEre_CAF1 95025 89 + 1 AGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAACGCGCUUUUUGCGAAUACGCUUCAUU-------CUUGU------------------------ .((.(((....))))).....(((((.......)))))(((((((((.....))))....(((.....))).....)))))...-------.....------------------------ ( -21.80) >DroMoj_CAF1 91616 115 + 1 AGUCGAUUUUUAUCACAGCGUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAUCGCGCUUUUUGCGGAUACGAUAUUUCUCGUCGUCUCUUUUUUUCUUUUCUUUUUGUU----- .(.(((....((((...((..(((((.......)))))...))((((.....))))(((.(((.....))))))..))))....))).)..........................----- ( -24.10) >DroAna_CAF1 230280 103 + 1 AGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAACGCGCUUUUUGCGGAUACGCUUUAUU-------CUCUUU--U--------ACUUAUUUGGAA .((.(((....)))))((((.(((((.......)))))(((((((((.....))))....(((.....))).....)))))...-------......--)--------)))......... ( -21.00) >consensus AGUCGAUUUUUAUCACAGUAUUUGAAUUAUUAUUUCAGGAAGCGCGCUAUUUGCGCAACGCGCUUUUUGCGGAUACGCUUUAUU_______CUCUUU__UU_______ACUUAUU_____ .((.(((....))))).(((((((...........(((((((((((............))))))))))))))))))............................................ (-17.89 = -17.95 + 0.06)



| Location | 15,363,104 – 15,363,207 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -13.05 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15363104 103 - 23771897 -----AAUAAGU---GGGGGA--AAAAAG-------AAUAAAGCGUAUCCGCAAAAAGCGCGUUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAAUACUGUGAUAAAAAUCGACU -----....(((---(.((((--(.....-------......(((....))).....((((((((......)))))))))))))(((.......)))...)))).((((....))))... ( -21.50) >DroPse_CAF1 96504 102 - 1 --AGAAAUAAGU-------AG--AAGGAG-------AAUAAAGCGUAUCCGCAAAAAGCGCGAUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAAUACUGUGAUAAAAAUCGACU --.......(((-------((--((((.(-------......((((((((((.....))).)))))))......).).)))).((((.......))))..)))).((((....))))... ( -21.00) >DroGri_CAF1 94140 108 - 1 -----GACAAAAAUAGAAAAAAAAAAAUG-------AAAAUAUUGUAUCCGCAAAAAGCGCGAUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAAUGCUGUGAUAAAAAUCGACU -----......................((-------((..((((((((((((.....))).)))((((.....))))..........)))))).)))).......((((....))))... ( -15.90) >DroEre_CAF1 95025 89 - 1 ------------------------ACAAG-------AAUGAAGCGUAUUCGCAAAAAGCGCGUUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAAUACUGUGAUAAAAAUCGACU ------------------------....(-------(((...(((....)))...((((((((((......))))))))))............))))........((((....))))... ( -19.40) >DroMoj_CAF1 91616 115 - 1 -----AACAAAAAGAAAAGAAAAAAAGAGACGACGAGAAAUAUCGUAUCCGCAAAAAGCGCGAUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAACGCUGUGAUAAAAAUCGACU -----............................(((....((((((((((((.....))).)))))....((((((.......((((.......))))..))))))))))....)))... ( -20.90) >DroAna_CAF1 230280 103 - 1 UUCCAAAUAAGU--------A--AAAGAG-------AAUAAAGCGUAUCCGCAAAAAGCGCGUUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAAUACUGUGAUAAAAAUCGACU ...((....(((--------(--...(((-------......(((....)))...((((((((((......)))))))))).............)))...)))).))............. ( -19.30) >consensus _____AAUAAGU_______AA__AAAAAG_______AAUAAAGCGUAUCCGCAAAAAGCGCGAUGCGCAAAUAGCGCGCUUCCUGAAAUAAUAAUUCAAAUACUGUGAUAAAAAUCGACU .............................................(((.((((..(((((((............)))))))..((((.......)))).....))))))).......... (-13.05 = -13.05 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:19 2006