| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,323,123 – 15,323,216 |
| Length | 93 |
| Max. P | 0.815846 |

| Location | 15,323,123 – 15,323,216 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15323123 93 + 23771897 U-------UUCCGCUUCGUGUUUGUUGCCAUUGUCCUGGAAAGUUUAGCGGCUUUAGC-CGCCUUGAUAAAACGACAGCCGCAACAAAA--ACAAG------CUGGAGA (-------((((((((.((.((((((((..(((((((....))....(((((....))-)))...........)))))..)))))))).--)))))------).))))) ( -35.10) >DroPse_CAF1 56836 100 + 1 UCAGUUAAUUCUGGUUGCUUUUUGUAGUC----UAGUGGAAAGUUUGGCGACUUUGGCUCCCCUUGAUACCGCUAUGGCCACAGCCCAAGGACA--CAACAGCCCG--- ............(((((....((((.(((----((((((((((((....))))))((....))......)))))..(((....)))...)))))--))))))))..--- ( -23.00) >DroSec_CAF1 55150 93 + 1 U-------UUCCGCUUCGUGUUUGUUGCCAUUGUCCUGGAAAGUUUAGCGGCUUUAGC-CGCCUUGAUAAAACGACAGCCGCAACAAAA--ACAAG------CUGGGAA .-------((((((((.((.((((((((..(((((((....))....(((((....))-)))...........)))))..)))))))).--)))))------).)))). ( -34.60) >DroSim_CAF1 55743 92 + 1 U-------UUCCGCUUCGUGUUUGUUGCCAUUGUCCUGGAAAGUUUAGCGGCU-UAGC-CGCCUUGAUAAAACGACAGCCGCAACAAAA--ACAAG------CUGGGAA .-------((((((((.((.((((((((..(((((((....))....(((((.-..))-)))...........)))))..)))))))).--)))))------).)))). ( -32.80) >DroEre_CAF1 55485 93 + 1 U-------UUCCGCUUCGUGUUUGUUGCCGUUGUCCUGGAAAGUUUAGCGGCUUUAGC-CGCCUUGAUAAAACGACAGCCGCAACAAAA--ACAAG------CUGGAAA .-------((((((((.((.((((((((.((((((((....))....(((((....))-)))...........)))))).)))))))).--)))))------).)))). ( -39.20) >DroYak_CAF1 57062 93 + 1 U-------UUCCGCUUCGUGUUUGUUGCCGUUGUCCUGGAAAGUUUAGCGGCUUUAGC-CGGCUUGAUAAAACGACAGCCGCAACAAAA--ACAAG------CUGGCAA .-------..((((((.((.((((((((.((((((((....))((.((((((....))-).))).))......)))))).)))))))).--)))))------).))... ( -33.10) >consensus U_______UUCCGCUUCGUGUUUGUUGCCAUUGUCCUGGAAAGUUUAGCGGCUUUAGC_CGCCUUGAUAAAACGACAGCCGCAACAAAA__ACAAG______CUGGAAA .................((.((((((((..(((((............(((((....)).)))...........)))))..))))))))...))................ (-13.11 = -14.08 + 0.97)

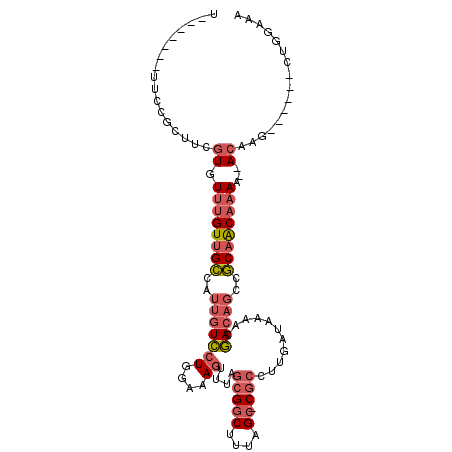
| Location | 15,323,123 – 15,323,216 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15323123 93 - 23771897 UCUCCAG------CUUGU--UUUUGUUGCGGCUGUCGUUUUAUCAAGGCG-GCUAAAGCCGCUAAACUUUCCAGGACAAUGGCAACAAACACGAAGCGGAA-------A ..(((.(------(((((--.((((((((.(.((((.(........((((-((....)))))).........).)))).).)))))))).)).))))))).-------. ( -35.13) >DroPse_CAF1 56836 100 - 1 ---CGGGCUGUUG--UGUCCUUGGGCUGUGGCCAUAGCGGUAUCAAGGGGAGCCAAAGUCGCCAAACUUUCCACUA----GACUACAAAAAGCAACCAGAAUUAACUGA ---...(((.(((--((((..((((((((....)))))(((..(..((....))...)..))).......)))...----))).))))..)))...(((......))). ( -22.80) >DroSec_CAF1 55150 93 - 1 UUCCCAG------CUUGU--UUUUGUUGCGGCUGUCGUUUUAUCAAGGCG-GCUAAAGCCGCUAAACUUUCCAGGACAAUGGCAACAAACACGAAGCGGAA-------A ((((..(------(((((--.((((((((.(.((((.(........((((-((....)))))).........).)))).).)))))))).)).))))))))-------. ( -34.53) >DroSim_CAF1 55743 92 - 1 UUCCCAG------CUUGU--UUUUGUUGCGGCUGUCGUUUUAUCAAGGCG-GCUA-AGCCGCUAAACUUUCCAGGACAAUGGCAACAAACACGAAGCGGAA-------A ((((..(------(((((--.((((((((.(.((((.(........((((-((..-.)))))).........).)))).).)))))))).)).))))))))-------. ( -32.73) >DroEre_CAF1 55485 93 - 1 UUUCCAG------CUUGU--UUUUGUUGCGGCUGUCGUUUUAUCAAGGCG-GCUAAAGCCGCUAAACUUUCCAGGACAACGGCAACAAACACGAAGCGGAA-------A .((((.(------(((((--.((((((((.(.((((.(........((((-((....)))))).........).)))).).)))))))).)).))))))))-------. ( -37.13) >DroYak_CAF1 57062 93 - 1 UUGCCAG------CUUGU--UUUUGUUGCGGCUGUCGUUUUAUCAAGCCG-GCUAAAGCCGCUAAACUUUCCAGGACAACGGCAACAAACACGAAGCGGAA-------A ...((.(------(((((--.((((((((.(.((((.........(((.(-((....))))))...((....)))))).).)))))))).)).))))))..-------. ( -30.80) >consensus UUCCCAG______CUUGU__UUUUGUUGCGGCUGUCGUUUUAUCAAGGCG_GCUAAAGCCGCUAAACUUUCCAGGACAAUGGCAACAAACACGAAGCGGAA_______A ..............((((...((((((((.(.(((((......)..((((.((....))))))...........)))).).)))))))).))))............... (-15.76 = -16.77 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:05 2006