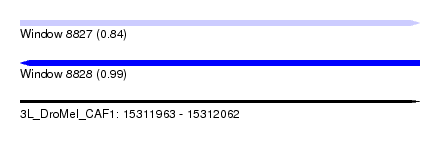
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,311,963 – 15,312,062 |
| Length | 99 |
| Max. P | 0.993549 |

| Location | 15,311,963 – 15,312,062 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -14.95 |
| Energy contribution | -16.48 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
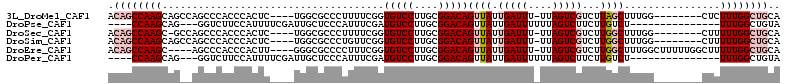
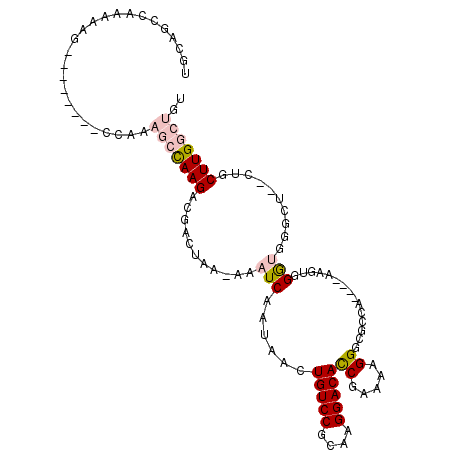
>3L_DroMel_CAF1 15311963 99 + 23771897 ACAGCCAAGCAGCCAGCCCACCCACUC----UGGCGCCCUUUUCGGUGUCCUUGCGGACAGUUAUUGAUUU-UUAGUCGUCUUAGCUUUGG--------CUCUUUGGCUGCA .((((((((.((((((.....((.(..----.((((((......))))))...).))..(((((.((((..-...))))...)))))))))--------)).)))))))).. ( -35.20) >DroPse_CAF1 46808 90 + 1 ----CCAAGCAG---GGUCUUCCAUUUUCGAUUGCUCCCAUUUCGAUGUCCUUGCGGACAGUUAUUGAUUUUUUAGUCUUCUUGUCU---------------UUUGGCUGUA ----((((((((---((.(........((((...........)))).).))))))((((((.....((((....))))...))))))---------------.))))..... ( -19.30) >DroSec_CAF1 43751 98 + 1 ACAGCCAAGC-GCCAGCCCACCCACUC----UGGCGCCCUUUUCGGUGUCCUUGCGGACAGUUAUUGAUUU-UUAGUCGUCUUGGCUUUGG--------CUUUUUGGCUGCA .((((((((.-(((((((.........----.((((((......)))))).....((((..(((.......-.)))..)))).)))..)))--------)..)))))))).. ( -32.30) >DroSim_CAF1 44385 99 + 1 ACAGCCAAGCAGCCAGCCCACCCACUC----UGGCGCCCUGUUCGGUGUCCUUGCGGACAGUUAUUGAUUU-UUAGUCGUCUUGGCUUUGG--------CUUUUUGGCUGCA .((((((((.((((((((.........----.)))((((((((((.........))))))).....(((..-...))).....)))..)))--------)).)))))))).. ( -35.80) >DroEre_CAF1 43698 103 + 1 ACAGCCAAGC----AGCCCACCCACUU----GGGCGCCCCUUUCGGUGUCCUUGCGGACAGUUAUUGAUUU-UUAGUCGUCUUGGCUUUGGCUUUUUGGCUUUUUGGCUGCA .((((((((.----((((...((.(..----(((((((......)))))))..).))..(((((.((((..-...))))...)))))..........)))).)))))))).. ( -37.70) >DroPer_CAF1 45549 90 + 1 ----CCAAGCAG---GGUCUUCCAUUUUCGAUUGCUCCCAUUUCGAUGUCCUUGCGGACAGUUAUUGAUUUUUUAGUCUUCUUGUCU---------------UUUGGCUGUA ----((((((((---((.(........((((...........)))).).))))))((((((.....((((....))))...))))))---------------.))))..... ( -19.30) >consensus ACAGCCAAGCAG__AGCCCACCCACUC____UGGCGCCCUUUUCGGUGUCCUUGCGGACAGUUAUUGAUUU_UUAGUCGUCUUGGCUUUGG________CUUUUUGGCUGCA .((((((((.....................................(((((....)))))((((.(((((....)))))...))))................)))))))).. (-14.95 = -16.48 + 1.53)
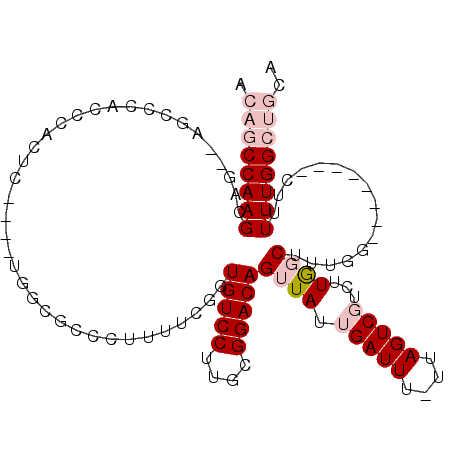
| Location | 15,311,963 – 15,312,062 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -17.43 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15311963 99 - 23771897 UGCAGCCAAAGAG--------CCAAAGCUAAGACGACUAA-AAAUCAAUAACUGUCCGCAAGGACACCGAAAAGGGCGCCA----GAGUGGGUGGGCUGGCUGCUUGGCUGU .((((((((..((--------(((..(((...........-...........(((((....)))))((.....)).((((.----.....))))))))))))..)))))))) ( -38.90) >DroPse_CAF1 46808 90 - 1 UACAGCCAAA---------------AGACAAGAAGACUAAAAAAUCAAUAACUGUCCGCAAGGACAUCGAAAUGGGAGCAAUCGAAAAUGGAAGACC---CUGCUUGG---- ..........---------------...((((.((.................(((((....)))))((((..((....)).))))............---)).)))).---- ( -17.80) >DroSec_CAF1 43751 98 - 1 UGCAGCCAAAAAG--------CCAAAGCCAAGACGACUAA-AAAUCAAUAACUGUCCGCAAGGACACCGAAAAGGGCGCCA----GAGUGGGUGGGCUGGC-GCUUGGCUGU .((((((((...(--------(((..(((...........-...........(((((....)))))((.....)).((((.----.....)))))))))))-..)))))))) ( -39.20) >DroSim_CAF1 44385 99 - 1 UGCAGCCAAAAAG--------CCAAAGCCAAGACGACUAA-AAAUCAAUAACUGUCCGCAAGGACACCGAACAGGGCGCCA----GAGUGGGUGGGCUGGCUGCUUGGCUGU .((((((((..((--------(((..(((...........-...........(((((....)))))((.....)).((((.----.....))))))))))))..)))))))) ( -41.60) >DroEre_CAF1 43698 103 - 1 UGCAGCCAAAAAGCCAAAAAGCCAAAGCCAAGACGACUAA-AAAUCAAUAACUGUCCGCAAGGACACCGAAAGGGGCGCCC----AAGUGGGUGGGCU----GCUUGGCUGU .((((((((..((((...........(((...........-...........(((((....)))))((....)))))((((----....)))).))))----..)))))))) ( -46.30) >DroPer_CAF1 45549 90 - 1 UACAGCCAAA---------------AGACAAGAAGACUAAAAAAUCAAUAACUGUCCGCAAGGACAUCGAAAUGGGAGCAAUCGAAAAUGGAAGACC---CUGCUUGG---- ..........---------------...((((.((.................(((((....)))))((((..((....)).))))............---)).)))).---- ( -17.80) >consensus UGCAGCCAAAAAG________CCAAAGCCAAGACGACUAA_AAAUCAAUAACUGUCCGCAAGGACACCGAAAAGGGCGCCA____AAGUGGGUGGGCU__CUGCUUGGCUGU .........................(((((((...........(((......(((((....)))))((.....))...............)))..........))))))).. (-17.43 = -18.18 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:56 2006