
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,309,921 – 15,310,032 |
| Length | 111 |
| Max. P | 0.846328 |

| Location | 15,309,921 – 15,310,032 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.50 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
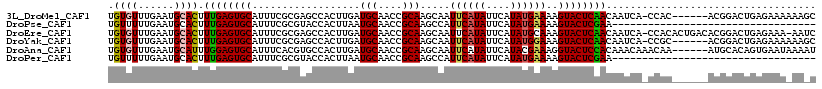
>3L_DroMel_CAF1 15309921 111 + 23771897 UGUGUUUGAAUGCACUUUGAGUGCAUUUCGCGAGCCACUUGAUGCAACCGCAAGCAAUUCAUAUUCAUAUGAAAAGUACUCAACAAUCA-CCAC------ACGGACUGAGAAAAAAGC .((((..((((((((.....))))))))...(((..((((..(((....))).....((((((....)))))))))).)))........-..))------))................ ( -22.70) >DroPse_CAF1 44758 84 + 1 UGUUUUUGAAUGCACUUUGAGUGCAUUUCGCGUACCACUUAAUGCAACCGCAAGCCAUUCAUAUUCAUAUGAAAAGUACUCGAA---------------------------------- ........(((((((.....)))))))(((.((((.......(((....))).....((((((....))))))..)))).))).---------------------------------- ( -20.40) >DroEre_CAF1 41755 116 + 1 UGUGUUUGAAUGCACUUUGAGUGCAUUUCGCGAGCCACUUGAUGCAACCGCAAGCAAUUCAUAUUCAUAUGCAAAGUACUCAACAAUCA-CCACACUGACACGGACUGAGAAA-AAUC .(((((.((((((((.....))))))))...(((..((((..(((....))).(((.............))).)))).)))........-.......)))))...........-.... ( -21.72) >DroYak_CAF1 39368 111 + 1 UGUGUUUGAAUGCACUUUGAGUGCAUUUCGCGAGCCACUUGAUGCAACCGCAAGCAAUUCAUAUUCAUAUGGAAAGUACUCAACAAUCA-CCGC------ACGGACUGAGAAAAAAGC .((((.(((.((....((((((((.(((((((.((........))...)))........((((....)))))))))))))))))).)))-..))------))................ ( -22.70) >DroAna_CAF1 39915 112 + 1 UGUGUUUGAAUGCAUUUGGAGUGCAUUUCACGUGCCACUUGAUGCAACCGCAAGCAAUUCAUAUUCAUACGAAAGGUACUCCACAAACAAACAA------AUGCACAGUGAAUAAAAU ((((((((..((....((((((((.((((..(((.....((((((........)))..)))....)))..)))).))))))))....))..)))------)))))............. ( -25.60) >DroPer_CAF1 43530 84 + 1 UGUUUUUGAAUGCACUUUGAGUGCAUUUCGCGUACCACUUAAUGCAACCGCAAGCCAUUCAUAUUCAUAUGAAAAGUACUCGAA---------------------------------- ........(((((((.....)))))))(((.((((.......(((....))).....((((((....))))))..)))).))).---------------------------------- ( -20.40) >consensus UGUGUUUGAAUGCACUUUGAGUGCAUUUCGCGAGCCACUUGAUGCAACCGCAAGCAAUUCAUAUUCAUAUGAAAAGUACUCAACAAUCA_CCAC______ACGGACUGAGAAA_AA_C .((((......)))).((((((((..................(((....))).....((((((....))))))..))))))))................................... (-16.52 = -16.50 + -0.02)

| Location | 15,309,921 – 15,310,032 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -19.43 |
| Energy contribution | -19.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15309921 111 - 23771897 GCUUUUUUCUCAGUCCGU------GUGG-UGAUUGUUGAGUACUUUUCAUAUGAAUAUGAAUUGCUUGCGGUUGCAUCAAGUGGCUCGCGAAAUGCACUCAAAGUGCAUUCAAACACA ................((------((..-(((.((((((((....((((((....))))))..(((((((((..(.....)..)).)))))...)))))))....))).))).)))). ( -29.10) >DroPse_CAF1 44758 84 - 1 ----------------------------------UUCGAGUACUUUUCAUAUGAAUAUGAAUGGCUUGCGGUUGCAUUAAGUGGUACGCGAAAUGCACUCAAAGUGCAUUCAAAAACA ----------------------------------.(((.(((((.((((((....))))))..((((((....)))...)))))))).)))(((((((.....)))))))........ ( -21.00) >DroEre_CAF1 41755 116 - 1 GAUU-UUUCUCAGUCCGUGUCAGUGUGG-UGAUUGUUGAGUACUUUGCAUAUGAAUAUGAAUUGCUUGCGGUUGCAUCAAGUGGCUCGCGAAAUGCACUCAAAGUGCAUUCAAACACA ....-.....((((.((((((((((..(-..(((....)))...)..))).)).))))).)))).(((((((..(.....)..)).)))))(((((((.....)))))))........ ( -27.30) >DroYak_CAF1 39368 111 - 1 GCUUUUUUCUCAGUCCGU------GCGG-UGAUUGUUGAGUACUUUCCAUAUGAAUAUGAAUUGCUUGCGGUUGCAUCAAGUGGCUCGCGAAAUGCACUCAAAGUGCAUUCAAACACA ................((------(...-(((.((((((((......((((....))))....(((((((((..(.....)..)).)))))...)))))))....))).)))..))). ( -24.70) >DroAna_CAF1 39915 112 - 1 AUUUUAUUCACUGUGCAU------UUGUUUGUUUGUGGAGUACCUUUCGUAUGAAUAUGAAUUGCUUGCGGUUGCAUCAAGUGGCACGUGAAAUGCACUCCAAAUGCAUUCAAACACA ............(((..(------(((..(((...((((((....((((((....))))))..(((..((((..(.....)..)).))..)...))))))))...)))..))))))). ( -29.50) >DroPer_CAF1 43530 84 - 1 ----------------------------------UUCGAGUACUUUUCAUAUGAAUAUGAAUGGCUUGCGGUUGCAUUAAGUGGUACGCGAAAUGCACUCAAAGUGCAUUCAAAAACA ----------------------------------.(((.(((((.((((((....))))))..((((((....)))...)))))))).)))(((((((.....)))))))........ ( -21.00) >consensus G_UU_UUUCUCAGUCCGU______GUGG_UGAUUGUUGAGUACUUUUCAUAUGAAUAUGAAUUGCUUGCGGUUGCAUCAAGUGGCACGCGAAAUGCACUCAAAGUGCAUUCAAACACA .....................................(((((...((((((....)))))).)))))(((((..(.....)..)).)))(((.(((((.....))))))))....... (-19.43 = -19.77 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:55 2006