| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
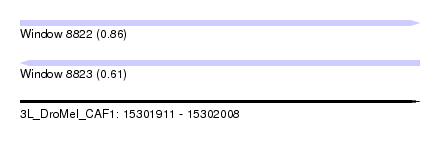
| Location | 15,301,911 – 15,302,008 |
| Length | 97 |
| Max. P | 0.864998 |

| Location | 15,301,911 – 15,302,008 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -19.78 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
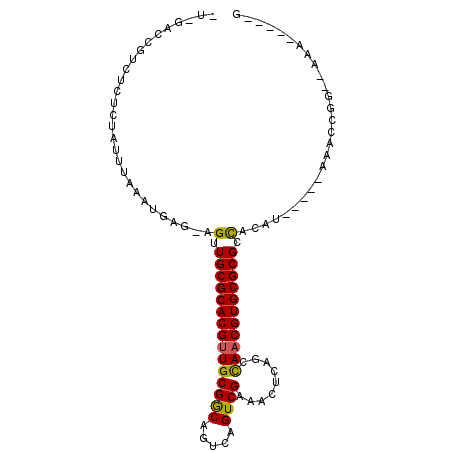
>3L_DroMel_CAF1 15301911 97 + 23771897 -U-GGACGACUCUCCAUUUAAAAGAGAAGUUGCGCACGUUGCGGCAGUCAGUCGAAAAUCAGCCAACGUGCGCGUUACAUUAC--GAACCGG--AAAAUAUCG -.-((..(((((((.........))).))))((((((((((((((.....)))).........))))))))))..........--...))..--......... ( -26.10) >DroVir_CAF1 34208 90 + 1 GUGUACCGUCUCUCUAUUUAAGUGAG-AGCUGCGCACGUUGCGGCAGUCAGUCGACACUCAGCCAACGUGCGCGCCAGAC-----AUACCAA--AAA-----G ((((....((((.((.....)).)))-)((.(((((((((((....(((....))).....).))))))))))))...))-----)).....--...-----. ( -29.00) >DroGri_CAF1 35294 87 + 1 ---GCCCGUCUCUCUAUUUAAAUGAG-AGUUGCGCACGUUGCGGCAGUCAGUCGACACUCAGCCAACGUGCGCGCCACAC-----AUACCGA--AAA-----G ---((....(((((.........)))-))..(((((((((((....(((....))).....).)))))))))))).....-----.......--...-----. ( -27.10) >DroSim_CAF1 34061 97 + 1 -U-GGACGACUCUCCAUUUAAAAGAGAAGUUGCGCACGUUGCGGCAGUCAGUCGAAAAUCAGCCAACGUGCGCGUUACAUUAC--GAACCGG--AAAGUAUCG -.-.((..(((.(((................((((((((((((((.....)))).........))))))))))((......))--.....))--).))).)). ( -26.60) >DroMoj_CAF1 32830 92 + 1 GAGGACCGUCUCUCUAUUUAAGUGAG-AGUUGCGCACGUUGCGGCAGUCAGUCGACACUCAGCUAACGUGCGCGCCACAU-----AUACCACAUAGA-----G ..((.....(((((.........)))-))..(((((((((((....(((....))).....)).))))))))).......-----...)).......-----. ( -28.60) >DroAna_CAF1 31985 87 + 1 -U-GU----------AUUUAAGUGAG-AGUUGCGCACGUUGCGACAGUCAGUCGAAAGUCAGCCAUCGUGCGCGCCACAUUACCGGAACCGG--A-AUUCUCG -.-..----------........(((-(((((((((((..(((((..((....))..))).))...))))))).........(((....)))--)-)))))). ( -32.20) >consensus _U_GACCGUCUCUCUAUUUAAAUGAG_AGUUGCGCACGUUGCGGCAGUCAGUCGAAACUCAGCCAACGUGCGCGCCACAU_____AAACCGG__AAA_____G ............................(.(((((((((((((((.....)))).........))))))))))).)........................... (-19.78 = -19.45 + -0.33)

| Location | 15,301,911 – 15,302,008 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15301911 97 - 23771897 CGAUAUUUU--CCGGUUC--GUAAUGUAACGCGCACGUUGGCUGAUUUUCGACUGACUGCCGCAACGUGCGCAACUUCUCUUUUAAAUGGAGAGUCGUCC-A- .((((((((--(((....--((......))((((((((((((................)))..)))))))))...............)))))))).))).-.- ( -27.79) >DroVir_CAF1 34208 90 - 1 C-----UUU--UUGGUAU-----GUCUGGCGCGCACGUUGGCUGAGUGUCGACUGACUGCCGCAACGUGCGCAGCU-CUCACUUAAAUAGAGAGACGGUACAC .-----...--...((((-----(((((((((((((((((.(.(...(((....)))..).))))))))))).)))-(((.........))))))).)))).. ( -31.50) >DroGri_CAF1 35294 87 - 1 C-----UUU--UCGGUAU-----GUGUGGCGCGCACGUUGGCUGAGUGUCGACUGACUGCCGCAACGUGCGCAACU-CUCAUUUAAAUAGAGAGACGGGC--- .-----..(--(((((((-----((((((((..((.((((((.....))))))))..)))))).))))))....((-(((.........))))).)))).--- ( -32.30) >DroSim_CAF1 34061 97 - 1 CGAUACUUU--CCGGUUC--GUAAUGUAACGCGCACGUUGGCUGAUUUUCGACUGACUGCCGCAACGUGCGCAACUUCUCUUUUAAAUGGAGAGUCGUCC-A- .((((((((--(((....--((......))((((((((((((................)))..)))))))))...............)))))))).))).-.- ( -30.09) >DroMoj_CAF1 32830 92 - 1 C-----UCUAUGUGGUAU-----AUGUGGCGCGCACGUUAGCUGAGUGUCGACUGACUGCCGCAACGUGCGCAACU-CUCACUUAAAUAGAGAGACGGUCCUC (-----(((((..(((..-----..((...(((((((((.((.(...(((....)))..).))))))))))).)).-...)))...))))))........... ( -30.00) >DroAna_CAF1 31985 87 - 1 CGAGAAU-U--CCGGUUCCGGUAAUGUGGCGCGCACGAUGGCUGACUUUCGACUGACUGUCGCAACGUGCGCAACU-CUCACUUAAAU----------AC-A- .((((((-(--(((....))).))).....(((((((...((.(((..((....))..)))))..)))))))...)-)))........----------..-.- ( -29.10) >consensus C_____UUU__CCGGUAC_____AUGUGGCGCGCACGUUGGCUGAGUGUCGACUGACUGCCGCAACGUGCGCAACU_CUCACUUAAAUAGAGAGACGGCC_A_ ..............................((((((((((((................)))..)))))))))............................... (-17.59 = -17.79 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:52 2006