
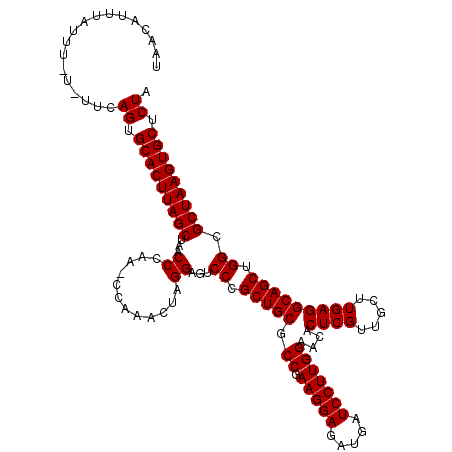
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,285,774 – 15,285,893 |
| Length | 119 |
| Max. P | 0.999899 |

| Location | 15,285,774 – 15,285,893 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -38.24 |
| Energy contribution | -38.24 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15285774 119 + 23771897 UAACAUUUAUUU-UGUUUAGUGCACUUAGCUAACCCAAACCAAACUAGGAGUCCCGCUGCGCCGAAGGAGAUGAUCCUUGGAACACUCGUUGCUUGAGGCAGCUGGCGCUAAGUGCUCUA .((((.......-))))(((.(((((((((.........((......)).(.((.(((((.((.(((((.....)))))))....((((.....))))))))).)))))))))))).))) ( -41.80) >DroSec_CAF1 17550 120 + 1 UAAUAUUUAUUUUUUUCCAGUGCACUUAGCUAGCCCAAUCCAAACUAGGAGUCCCGCUGCGCCGAAGGAGAUGAUCCUUGGAACACUCGUUGCUUGAGGCAGCUGGCGCUAAGUGCUCUA ..................((.(((((((((..(((...(((......))).....(((((.((.(((((.....)))))))....((((.....))))))))).)))))))))))).)). ( -42.80) >DroSim_CAF1 17635 119 + 1 UAACAUUUAUUU-UGUCCAGUGCACUUAGCUAGCCCAAACCAAACUAGGAGUCCCGCUGCGCCGAAGGAGAUGAUCCUUGGAACACUCGUUGCUUGAGGCAGCUGGCGCUAAGUGCUCUA ............-.....((.(((((((((..(((............((....))(((((.((.(((((.....)))))))....((((.....))))))))).)))))))))))).)). ( -41.50) >DroEre_CAF1 17547 113 + 1 UAACAUUCCUUU-U-CUCAGUGCACUUAGCUAACCC-----AAACUGGGAAUCCCGCUGCGCCGAAGGAGCUGGUCCUUGGAAUACUCGAUGCUUGAGGCAGCUGGCGCUAAGUGCUCUA ............-.-...((.(((((((((...(((-----.....)))...((.(((((.((.(((((.....)))))))....(((((...)))))))))).)).))))))))).)). ( -42.20) >DroYak_CAF1 14606 113 + 1 UUAUACUUAUUC-U-UUCAGUGCACUUAGCUAACCC-----AAACUGGGAAUCCCGCUGCGCCGAAGGAGAUGGUCCUUGGAAUACUCGAUGCUUGAGGCAGCUGGCGCUAAGUGCUCUA ............-.-...((.(((((((((...(((-----.....)))...((.(((((.((.(((((.....)))))))....(((((...)))))))))).)).))))))))).)). ( -42.20) >consensus UAACAUUUAUUU_U_UUCAGUGCACUUAGCUAACCCAA_CCAAACUAGGAGUCCCGCUGCGCCGAAGGAGAUGAUCCUUGGAACACUCGUUGCUUGAGGCAGCUGGCGCUAAGUGCUCUA ..................((.(((((((((...((............))...((.(((((.((.(((((.....)))))))....((((.....))))))))).)).))))))))).)). (-38.24 = -38.24 + -0.00)


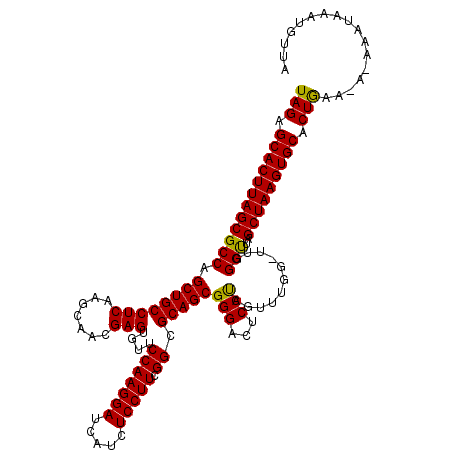
| Location | 15,285,774 – 15,285,893 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -38.50 |
| Energy contribution | -37.86 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15285774 119 - 23771897 UAGAGCACUUAGCGCCAGCUGCCUCAAGCAACGAGUGUUCCAAGGAUCAUCUCCUUCGGCGCAGCGGGACUCCUAGUUUGGUUUGGGUUAGCUAAGUGCACUAAACA-AAAUAAAUGUUA (((.(((((((((.((.(((((((.(((....(((((.((....)).)).)))))).)).))))).))...(((((......)))))...))))))))).)))((((-.......)))). ( -39.80) >DroSec_CAF1 17550 120 - 1 UAGAGCACUUAGCGCCAGCUGCCUCAAGCAACGAGUGUUCCAAGGAUCAUCUCCUUCGGCGCAGCGGGACUCCUAGUUUGGAUUGGGCUAGCUAAGUGCACUGGAAAAAAAUAAAUAUUA (((.((((((((((((.(((((((.(((....(((((.((....)).)).)))))).)).))))).....(((......)))...)))..))))))))).)))................. ( -39.90) >DroSim_CAF1 17635 119 - 1 UAGAGCACUUAGCGCCAGCUGCCUCAAGCAACGAGUGUUCCAAGGAUCAUCUCCUUCGGCGCAGCGGGACUCCUAGUUUGGUUUGGGCUAGCUAAGUGCACUGGACA-AAAUAAAUGUUA (((.(((((((((.((.(((((((.(((....(((((.((....)).)).)))))).)).))))).))...(((((......)))))...))))))))).)))((((-.......)))). ( -39.80) >DroEre_CAF1 17547 113 - 1 UAGAGCACUUAGCGCCAGCUGCCUCAAGCAUCGAGUAUUCCAAGGACCAGCUCCUUCGGCGCAGCGGGAUUCCCAGUUU-----GGGUUAGCUAAGUGCACUGAG-A-AAAGGAAUGUUA (((.(((((((((.((.((((((((.......)))....(((((((.....))))).)).))))).))...(((.....-----)))...))))))))).)))..-.-............ ( -39.40) >DroYak_CAF1 14606 113 - 1 UAGAGCACUUAGCGCCAGCUGCCUCAAGCAUCGAGUAUUCCAAGGACCAUCUCCUUCGGCGCAGCGGGAUUCCCAGUUU-----GGGUUAGCUAAGUGCACUGAA-A-GAAUAAGUAUAA (((.(((((((((.((.((((((((.......)))....(((((((.....))))).)).))))).))...(((.....-----)))...))))))))).)))..-.-............ ( -39.40) >consensus UAGAGCACUUAGCGCCAGCUGCCUCAAGCAACGAGUGUUCCAAGGAUCAUCUCCUUCGGCGCAGCGGGACUCCUAGUUUGG_UUGGGUUAGCUAAGUGCACUGAA_A_AAAUAAAUGUUA (((.((((((((((((.((((((((.......)))....(((((((.....))))).)).)))))(((...)))...........)))..))))))))).)))................. (-38.50 = -37.86 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:45 2006