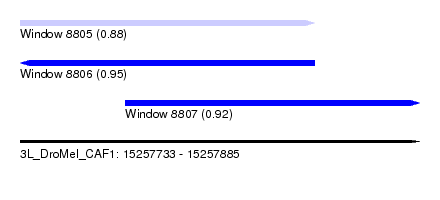
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,257,733 – 15,257,885 |
| Length | 152 |
| Max. P | 0.953579 |

| Location | 15,257,733 – 15,257,845 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.48 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15257733 112 + 23771897 UCGAAGUUUACUACUCAUUCUUCUUUAUUAUUCUUGAUUGUUCCCAACAAAGCUUUGAAAACCAUCAUACCCCAUUGGCCGUAACGGCCCCAGGGUAUGUAACGAACCGAAU (((((((((.....(((.................))).(((.....)))))))))))).......(((((((....(((((...)))))...)))))))............. ( -24.53) >DroSec_CAF1 146910 107 + 1 UUAAAGUUUAGUACUAAUUUUUUUUUA----UCUUGUUUAUUCCCAACGUUGAA-UAAAAAUCAUCAUACCCCAUUUGCCUUAAAGGCUCCAGGGUAUGUAACGAACCGAAU .....((((..(((.............----.....(((((((........)))-)))).......((((((.....(((.....)))....)))))))))..))))..... ( -19.50) >DroSim_CAF1 155140 112 + 1 UUAAAGUUUACUACGCAUUCUUUUUUAAUAUUCUUGUUUAUUCCCAACGUAGCUUUGAAAACCAUCAUACCCCAUUGGCCUUAAAGGCCCCAGGGUAUGUAACGAACCGAAU ((((((....(((((...........((((....)))).........))))))))))).......(((((((....((((.....))))...)))))))............. ( -22.85) >DroEre_CAF1 147174 109 + 1 CUAAAUUCC---UCUGAUUCUUUUUUAUUACUCUUGCACAUUCCAAACGAUGCUGUGAAAACCAUCAUGCCCCAUUGGCCGUAAAAGCCACAGGGUAUGUAACGAACCGAAU .........---....((((...((..((((.................((((..((....))))))((((((...((((.......))))..))))))))))..))..)))) ( -20.40) >DroYak_CAF1 153365 107 + 1 CUAAAGUUC-----ACAUUCUUUCGCAUUUUUCUUUUGCAUUCCAAGCGAUGCCUUGAAAAACAUCAUACCCCAGUGGCCCUAGAGGCCCCAGGGUAUGUAACGAACCGAAU .....((((-----.............((((((....(((((......)))))...))))))...(((((((..(.((((.....)))))..)))))))....))))..... ( -28.60) >consensus UUAAAGUUUA_UACUCAUUCUUUUUUAUUAUUCUUGUUUAUUCCCAACGAUGCUUUGAAAACCAUCAUACCCCAUUGGCCGUAAAGGCCCCAGGGUAUGUAACGAACCGAAU .................................................................(((((((....((((.....))))...)))))))............. (-15.24 = -15.48 + 0.24)


| Location | 15,257,733 – 15,257,845 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.78 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15257733 112 - 23771897 AUUCGGUUCGUUACAUACCCUGGGGCCGUUACGGCCAAUGGGGUAUGAUGGUUUUCAAAGCUUUGUUGGGAACAAUCAAGAAUAAUAAAGAAGAAUGAGUAGUAAACUUCGA ....((((..(((((((((((..(((((...)))))...)))))))(((.(((..(((.......)))..))).))).....................))))..)))).... ( -33.30) >DroSec_CAF1 146910 107 - 1 AUUCGGUUCGUUACAUACCCUGGAGCCUUUAAGGCAAAUGGGGUAUGAUGAUUUUUA-UUCAACGUUGGGAAUAAACAAGA----UAAAAAAAAAUUAGUACUAAACUUUAA ....(((.((((((((((((((..(((.....)))...))))))))).)))).((((-(((........))))))).....----.............).)))......... ( -22.50) >DroSim_CAF1 155140 112 - 1 AUUCGGUUCGUUACAUACCCUGGGGCCUUUAAGGCCAAUGGGGUAUGAUGGUUUUCAAAGCUACGUUGGGAAUAAACAAGAAUAUUAAAAAAGAAUGCGUAGUAAACUUUAA ....((((..(((((((((((..((((.....))))...)))))))....(((..(((.......)))..))).........................))))..)))).... ( -28.50) >DroEre_CAF1 147174 109 - 1 AUUCGGUUCGUUACAUACCCUGUGGCUUUUACGGCCAAUGGGGCAUGAUGGUUUUCACAGCAUCGUUUGGAAUGUGCAAGAGUAAUAAAAAAGAAUCAGA---GGAAUUUAG .((((((((....(((.((((((((((.....)))).)))))).))).((.((((((((...((.....)).)))).)))).))........))))).))---)........ ( -26.00) >DroYak_CAF1 153365 107 - 1 AUUCGGUUCGUUACAUACCCUGGGGCCUCUAGGGCCACUGGGGUAUGAUGUUUUUCAAGGCAUCGCUUGGAAUGCAAAAGAAAAAUGCGAAAGAAUGU-----GAACUUUAG ((((..(((((..(((((((..(((((.....)))).)..))))))).(((.(((((((......))))))).)))..........))))).))))..-----......... ( -39.90) >consensus AUUCGGUUCGUUACAUACCCUGGGGCCUUUAAGGCCAAUGGGGUAUGAUGGUUUUCAAAGCAUCGUUGGGAAUAAACAAGAAUAAUAAAAAAGAAUGAGUA_UAAACUUUAA .....((((....((((((((..((((.....))))...)))))))).............((.....))))))....................................... (-19.82 = -19.78 + -0.04)

| Location | 15,257,773 – 15,257,885 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15257773 112 + 23771897 UUCCCAACAAAGCUUUGAAAACCAUCAUACCCCAUU----GGCCGUAACGG----CCCCAGGGUAUGUAACGAACCGAAUUACAAGGCGUACAACCCUCAGGACAACUGUCCAACUUGGA ...((((....((((((........(((((((....----(((((...)))----))...)))))))...((...)).....))))))............((((....))))...)))). ( -33.50) >DroSec_CAF1 146946 111 + 1 UUCCCAACGUUGAA-UAAAAAUCAUCAUACCCCAUU----UGCCUUAAAGG----CUCCAGGGUAUGUAACGAACCGAAUUACAAGGCGUACAACCCUCAGGACAACUGUCCAACUUGGA ...((((.((((..-..........(((((((....----.(((.....))----)....)))))))..(((..((.........))))).)))).....((((....))))...)))). ( -28.50) >DroSim_CAF1 155180 112 + 1 UUCCCAACGUAGCUUUGAAAACCAUCAUACCCCAUU----GGCCUUAAAGG----CCCCAGGGUAUGUAACGAACCGAAUUACAAGGCGUACAACCCUCAGGACAACUGUCCAACUUGGA ...((((.(((((((((........(((((((....----((((.....))----))...)))))))...((...)).....)))))).)))........((((....))))...)))). ( -34.80) >DroEre_CAF1 147211 112 + 1 UUCCAAACGAUGCUGUGAAAACCAUCAUGCCCCAUU----GGCCGUAAAAG----CCACAGGGUAUGUAACGAACCGAAUUACAAGGCGUACAACCCUCAGGACAACUGUCCAACUUGGA .(((((...(((((((....))...(((((((...(----(((.......)----)))..)))))))..................)))))..........((((....))))...))))) ( -30.20) >DroYak_CAF1 153400 112 + 1 UUCCAAGCGAUGCCUUGAAAAACAUCAUACCCCAGU----GGCCCUAGAGG----CCCCAGGGUAUGUAACGAACCGAAUUACAAGGCGUACAACCCUCAGGACAACUGUCCAACUUGGC ..(((((..((((((((........(((((((..(.----((((.....))----)))..)))))))...((...)).....))))))))..........((((....))))..))))). ( -39.50) >DroAna_CAF1 139208 104 + 1 ---------------UAAAAAAUAAUAUCCCUCAGUUUAUGGCCA-AUACACCUUUUCUAGGGUAAAAAACGUAACGAAUUACAAGGAGUACACCCCUUAGGACAACUGUCCAAUUUGGA ---------------.....................(((((....-.(((.(((.....)))))).....)))))((((((..((((.(....).)))).((((....)))))))))).. ( -18.80) >consensus UUCCCAACGAUGCUUUGAAAACCAUCAUACCCCAUU____GGCCGUAAAGG____CCCCAGGGUAUGUAACGAACCGAAUUACAAGGCGUACAACCCUCAGGACAACUGUCCAACUUGGA .........................(((((((............................))))))).......((((......(((.(.....))))..((((....))))...)))). (-15.30 = -15.27 + -0.02)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:36 2006