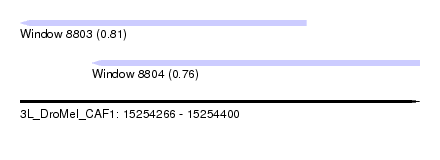
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,254,266 – 15,254,400 |
| Length | 134 |
| Max. P | 0.814052 |

| Location | 15,254,266 – 15,254,362 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15254266 96 - 23771897 CAGUAUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG--------GAAUUGAAAACUUCAACAAGUAUUCU--- .((((((((((((((((((((((.....))))))))).))))))))))))).......((((((......--------(((.(....).)))......))))))--- ( -26.50) >DroSec_CAF1 143447 92 - 1 ----AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG--------GAAUUGAAAACUUCAACAAGUAUUCU--- ----.....(((((......)).)))((((((((((.(((((((((.(.((.((...........)).))--------.).)))))))))..))))))))))..--- ( -24.30) >DroSim_CAF1 151659 92 - 1 ----AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG--------GAAUUGAGAACUUCAACAAGUAUUCU--- ----.....(((((......)).)))((((((((((.(((((((((.(.((.((...........)).))--------.).)))))))))..))))))))))..--- ( -24.40) >DroEre_CAF1 143802 103 - 1 ----AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUGGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAGGAACCAAGGAACUGAAAACUUCUACAAGUAUUCUUCU ----.....(((((......)).)))(((((((((.(((((((((..(.((.((..(..((....))..)...)).)).)..)))))))))..)))))))))..... ( -26.70) >DroYak_CAF1 149855 95 - 1 ----AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUGGGUUUUCAAGUGCUCGGAACAAGGAUAACCAAA--------GAAUGGAAAACUUCAACAAGUAUUCUUCU ----.....(((((......)).)))((((((((((((((((((..((.......))..((....))...--------.....)))))).))))))))))))..... ( -23.80) >consensus ____AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG________GAAUUGAAAACUUCAACAAGUAUUCU___ .........(((((......)).)))(((((((((..(((((((((((.......)).((.....))..............)))))))))...)))))))))..... (-20.18 = -20.62 + 0.44)

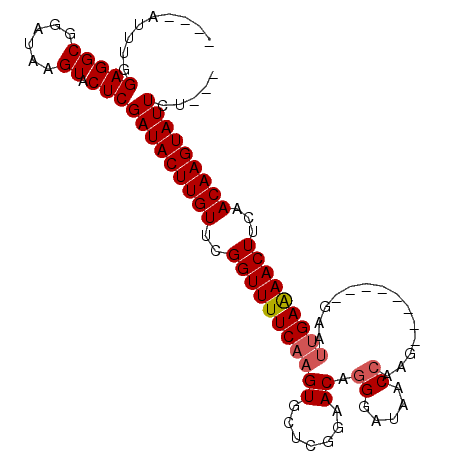

| Location | 15,254,290 – 15,254,400 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -19.85 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15254290 110 - 23771897 GUAAUAAAGAA--UAACAAGGAUAAUCCAGUGAAAGUAUCCAGUAUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG--------GA ...........--............(((.......((.(((((((((((((((((((((((((.....))))))))).))))))))))))).))))).((.....))..)--------)) ( -31.80) >DroSec_CAF1 143471 105 - 1 GUAAUAAAGAAUAUAACAAGGAUAAUCCGGUGAAAGU-------AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG--------GA ...................((...((((.((...(((-------(((((((((((((((((((.....))))))))).)))))))))))))....))..))))..))...--------.. ( -29.20) >DroSim_CAF1 151683 103 - 1 GUAAUAAAGAA--UAACAAGGAUAAUCCAGUGAAAGU-------AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG--------GA ...........--......((.((.(((.((...(((-------(((((((((((((((((((.....))))))))).)))))))))))))....)).))).)).))...--------.. ( -28.40) >DroEre_CAF1 143829 111 - 1 GUAAUAAAGAA--UAACAAGGAUAAUCCAGUGAAAGU-------AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUGGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAGGAACCAAGGA ...........--......((....(((.((...(((-------((((((((((.((((((((.....)))))))).).))))))))))))....)).((.....))..))).))..... ( -26.20) >DroYak_CAF1 149882 103 - 1 GUAAUAAGGAA--UAACAAGGAUAAUCCAGUGAAAGU-------AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUGGGUUUUCAAGUGCUCGGAACAAGGAUAACCAAA--------GA ...........--......((...((((.((...(((-------((((((((((.((((((((.....)))))))).).))))))))))))....))..))))..))...--------.. ( -24.70) >consensus GUAAUAAAGAA__UAACAAGGAUAAUCCAGUGAAAGU_______AUUUGGAGGCGGAUAAGUACUCGAUACUUGUUCGGUUUUCAAGUGCUCGGAACAGGGAUAACCAAG________GA ...................((...((((.((...(((.......(((((((((((((((((((.....))))))))).)))))))))))))....))..))))..))............. (-19.85 = -20.25 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:33 2006