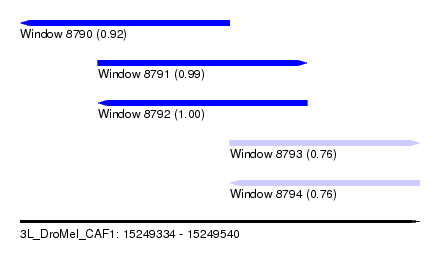
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,249,334 – 15,249,540 |
| Length | 206 |
| Max. P | 0.999988 |

| Location | 15,249,334 – 15,249,442 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -22.05 |
| Energy contribution | -23.05 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15249334 108 - 23771897 UUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAUUCGGAUUCGGAUUCUGGCACAUCACAUCUUAAGCUUCAAUUAUCACCGCCGUAUUAUUGCAUUUUCACAUGGU ..((.(((((.(((((.((((((...------------..)))))).))))).))))).)).(((.............................)))(((....)))............. ( -22.95) >DroSec_CAF1 138516 102 - 1 UUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAU------UCGUAUUCUGGCACAUCACAUCUUAAGCUUCAAUUAUCACCGCCGUCUUAUUGCAUUUGCACAUGAU ..(((...((...(((.((((((...------------..)))))).------)))...)).(((.............................))))))(((((((....))).)))). ( -17.85) >DroSim_CAF1 146261 108 - 1 UUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAUUCAGAUUCGGAUUCUGGCACAUCACAUCUUAAGCUUCAAUUAUCACCGCCGUCUUAUUGCAUUUGCACAUGAU ..((((((((.(((((.((((((...------------..)))))).))))).)))))....(((.............................))))))(((((((....))).)))). ( -26.15) >DroEre_CAF1 138854 120 - 1 UUGACUCUGAUUUUGAUUCCGAUUCCGAUUCCGCUGCCGAAUCGGAUUCGUAUUCGGAUUCAGGCACAUCACAUCUUAAGCUUCAAUUAUCACCGCCGUCUUGUUGCAUUUGCACAUGGU ((((..((((.(((((...(((.((((((((.......)))))))).)))...))))).))))((..............)).))))........((((...(((.((....))))))))) ( -29.24) >DroYak_CAF1 144789 108 - 1 UUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAUUCGGAUUCGGAUUCAGGCACAUCACAUCUUAAGCUUCAAUUAUCACCGCCGUCUUGUUGCAUUUGCACAUGGU ((((..((((.(((((.(((((.(((------------(...)))).))))).))))).))))((..............)).))))........((((...(((.((....))))))))) ( -28.24) >consensus UUGACUCUGAUUUUGAUUCCGAUUCC____________GAAUCGGAUUCGGAUUCGGAUUCUGGCACAUCACAUCUUAAGCUUCAAUUAUCACCGCCGUCUUAUUGCAUUUGCACAUGGU ..((.(((((.(((((.((((((((.............)))))))).))))).))))).)).(((.............................)))...(((((((....))).)))). (-22.05 = -23.05 + 1.00)

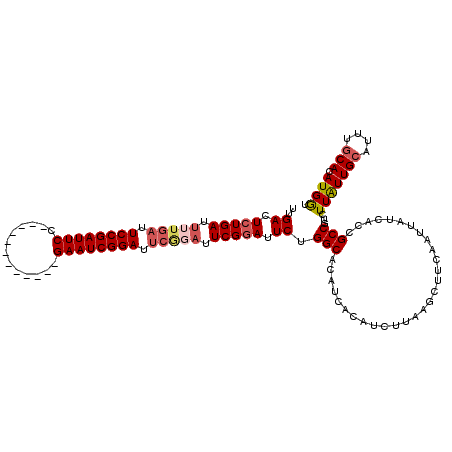
| Location | 15,249,374 – 15,249,482 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -25.72 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15249374 108 + 23771897 CUUAAGAUGUGAUGUGCCAGAAUCCGAAUCCGAAUCCGAUUC------------GGAAUCGGAAUCAAAAUCAGAGUCAAAGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUA ......((((((((.((..((.(((((.(((((((...))))------------))).))))).)).........(.((((....)))).))))))..(((((....)))))..))))). ( -32.20) >DroSec_CAF1 138556 102 + 1 CUUAAGAUGUGAUGUGCCAGAAUACGA------AUCCGAUUC------------GGAAUCGGAAUCAAAAUCAGAGUCAAAGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUA ......(((((..(((.((((....((------.((((((..------------...)))))).))........(((....))).)))).))).....(((((....)))))..))))). ( -24.10) >DroSim_CAF1 146301 108 + 1 CUUAAGAUGUGAUGUGCCAGAAUCCGAAUCUGAAUCCGAUUC------------GGAAUCGGAAUCAAAAUCAGAGUCAAAGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUA ......((((((((.((..((.(((((.(((((((...))))------------))).))))).)).........(.((((....)))).))))))..(((((....)))))..))))). ( -30.00) >DroEre_CAF1 138894 120 + 1 CUUAAGAUGUGAUGUGCCUGAAUCCGAAUACGAAUCCGAUUCGGCAGCGGAAUCGGAAUCGGAAUCAAAAUCAGAGUCAAAGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUA ......((((((((.((((((....((...(((.((((((((.......)))))))).)))...))....)))).(.((((....)))).))))))..(((((....)))))..))))). ( -34.00) >DroYak_CAF1 144829 108 + 1 CUUAAGAUGUGAUGUGCCUGAAUCCGAAUCCGAAUCCGAUUC------------GGAAUCGGAAUCAAAAUCAGAGUCAAAGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUA ......((((((((.((.(((.(((((.(((((((...))))------------))).))))).)))........(.((((....)))).))))))..(((((....)))))..))))). ( -32.80) >consensus CUUAAGAUGUGAUGUGCCAGAAUCCGAAUCCGAAUCCGAUUC____________GGAAUCGGAAUCAAAAUCAGAGUCAAAGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUA ......((((((((.((..((.((.((....((.((((((((.............)))))))).))....)).)).))...((.....)).)))))..(((((....)))))..))))). (-25.72 = -26.32 + 0.60)

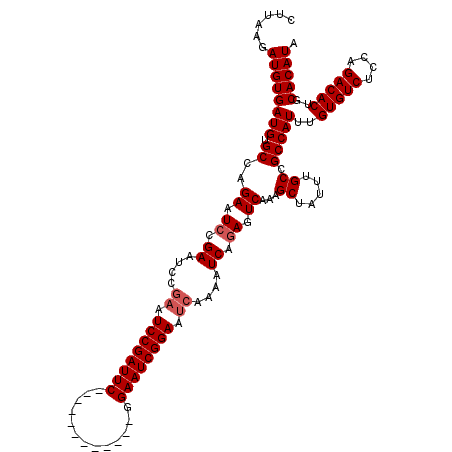

| Location | 15,249,374 – 15,249,482 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -35.12 |
| Energy contribution | -36.40 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.48 |
| SVM RNA-class probability | 0.999988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15249374 108 - 23771897 UAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCUUUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAUUCGGAUUCGGAUUCUGGCACAUCACAUCUUAAG .((((((.(((((....)))))....(((........)))..((.(((((.(((((.((((((...------------..)))))).))))).))))).))..))))))........... ( -35.90) >DroSec_CAF1 138556 102 - 1 UAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCUUUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAU------UCGUAUUCUGGCACAUCACAUCUUAAG .((((((.(((((....))))).....(((((.....)))..((.((((((((.((.......)).------------)))))))).------))))......))))))........... ( -27.00) >DroSim_CAF1 146301 108 - 1 UAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCUUUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAUUCAGAUUCGGAUUCUGGCACAUCACAUCUUAAG .((((((.(((((....)))))....(((........)))..((.(((((.(((((.((((((...------------..)))))).))))).))))).))..))))))........... ( -36.70) >DroEre_CAF1 138894 120 - 1 UAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCUUUGACUCUGAUUUUGAUUCCGAUUCCGAUUCCGCUGCCGAAUCGGAUUCGUAUUCGGAUUCAGGCACAUCACAUCUUAAG .((((((.(((((....)))))....(((........)))......((((.(((((...(((.((((((((.......)))))))).)))...))))).))))))))))........... ( -40.10) >DroYak_CAF1 144829 108 - 1 UAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCUUUGACUCUGAUUUUGAUUCCGAUUCC------------GAAUCGGAUUCGGAUUCGGAUUCAGGCACAUCACAUCUUAAG .((((((.(((((....)))))....(((........)))......((((.(((((.(((((.(((------------(...)))).))))).))))).))))))))))........... ( -39.10) >consensus UAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCUUUGACUCUGAUUUUGAUUCCGAUUCC____________GAAUCGGAUUCGGAUUCGGAUUCUGGCACAUCACAUCUUAAG .((((((.(((((....)))))....(((........)))..((.(((((.(((((.((((((((.............)))))))).))))).))))).))..))))))........... (-35.12 = -36.40 + 1.28)


| Location | 15,249,442 – 15,249,540 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -24.73 |
| Energy contribution | -25.90 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15249442 98 + 23771897 AGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUAUGACAUUGUGCAUCACCGGCAGGACAAAGGCCAAGAAGAGUGGGCGGCAA----------------------GUGGGCGG ..(((((((((((((((((((((....)))))((((((........)))))).....(((.........))).....)))).))))))))----------------------)))).... ( -40.00) >DroSec_CAF1 138618 98 + 1 AGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUAUGACAUUGUGCAUCACCGGCAGGACAAAGGCCAAGCAGAGUGGGCGGCAA----------------------GUGGGCGG ..((((((((((((....(((((....)))))((((((........)))))).((((.((.((.......))..)).).)))))))))))----------------------)))).... ( -40.40) >DroSim_CAF1 146369 98 + 1 AGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUAUGACAUUGUGCAUCACUGGCAGGACAAAGGCCAAGCAGAGUGGGCGGCAA----------------------GUGGGCGG ..((((((((((((....(((((....)))))((((((........)))))).((((.((.((.......))..))..))))))))))))----------------------)))).... ( -41.80) >DroEre_CAF1 138974 96 + 1 AGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUAUGACAUUGUGCAUCAGCGGCAGGACAAAGGCCAAGAAGAGUGGGCG-----------------------GCAGUGGGCG- .(((..(((((((((((((((((....)))))((((((........)))))).....(((.........))).....)))).))))-----------------------))))..))).- ( -38.10) >DroYak_CAF1 144897 119 + 1 AGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUAUGACAUUGUGCAUCAGCGGCAGGACAAAGGCCAAGAAGAGUGGGCGCAAAGUGGGCGUGGAGUGUGCGGGGAGUGGGCA- ..((((((.((((((((((((((....)))))((((((........))))))...(((.(........(.(((.......))).)........).))).))))).)))).))))))...- ( -40.89) >DroAna_CAF1 131119 84 + 1 AGCUAUUUGCCACCAUUUCUGUCUCCAGACACAGCACAUAUGACAAUGUGCAUCAGCGGCAGGACAAAGGCCAGGAAAAGCGGA------------------------------------ .(((...((((........((((....))))..((((((......))))))......))))((.......))......)))...------------------------------------ ( -21.40) >consensus AGCUAUUUGCCGCCAUUUGUGUCUCCAGACACUGCACAUAUGACAUUGUGCAUCACCGGCAGGACAAAGGCCAAGAAGAGUGGGCGGCAA______________________GUGGGCG_ .(((.(((((((((....(((((....))))).(((((........)))))......))).)).))))(.(((.......))).)..............................))).. (-24.73 = -25.90 + 1.17)


| Location | 15,249,442 – 15,249,540 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -23.79 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15249442 98 - 23771897 CCGCCCAC----------------------UUGCCGCCCACUCUUCUUGGCCUUUGUCCUGCCGGUGAUGCACAAUGUCAUAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCU ..((....----------------------((((((((((((......(((.........))))))).((((((........))))))(((((....)))))....))))))))...)). ( -34.20) >DroSec_CAF1 138618 98 - 1 CCGCCCAC----------------------UUGCCGCCCACUCUGCUUGGCCUUUGUCCUGCCGGUGAUGCACAAUGUCAUAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCU ..((....----------------------((((((((((((..((..(((....)))..)).)))).((((((........))))))(((((....)))))....))))))))...)). ( -37.10) >DroSim_CAF1 146369 98 - 1 CCGCCCAC----------------------UUGCCGCCCACUCUGCUUGGCCUUUGUCCUGCCAGUGAUGCACAAUGUCAUAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCU ..((....----------------------((((((((((((..((..(((....)))..)).)))).((((((........))))))(((((....)))))....))))))))...)). ( -37.10) >DroEre_CAF1 138974 96 - 1 -CGCCCACUGC-----------------------CGCCCACUCUUCUUGGCCUUUGUCCUGCCGCUGAUGCACAAUGUCAUAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCU -.((....(((-----------------------((((......((.((((.........))))..))((((((........))))))(((((....)))))....)))))))....)). ( -32.80) >DroYak_CAF1 144897 119 - 1 -UGCCCACUCCCCGCACACUCCACGCCCACUUUGCGCCCACUCUUCUUGGCCUUUGUCCUGCCGCUGAUGCACAAUGUCAUAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCU -............(((((......((.......))(((..........)))...)))..(((((((..((((((........))))))(((((....)))))....)))))))....)). ( -31.40) >DroAna_CAF1 131119 84 - 1 ------------------------------------UCCGCUUUUCCUGGCCUUUGUCCUGCCGCUGAUGCACAUUGUCAUAUGUGCUGUGUCUGGAGACAGAAAUGGUGGCAAAUAGCU ------------------------------------...(((......(((....))).(((((((...((((((......))))))....((((....))))...)))))))...))). ( -27.60) >consensus _CGCCCAC______________________UUGCCGCCCACUCUUCUUGGCCUUUGUCCUGCCGCUGAUGCACAAUGUCAUAUGUGCAGUGUCUGGAGACACAAAUGGCGGCAAAUAGCU ................................................(((.((((.((.((((....((((((........))))))(((((....)))))...))))))))))..))) (-23.79 = -23.85 + 0.06)

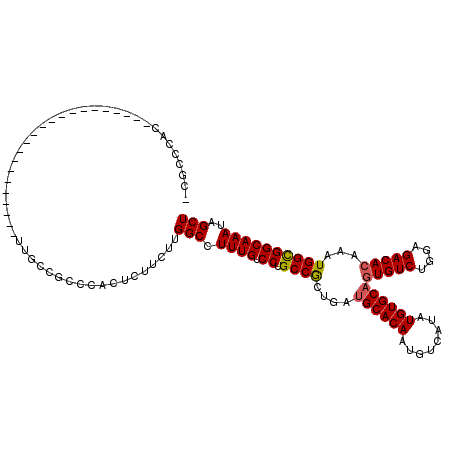
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:24 2006