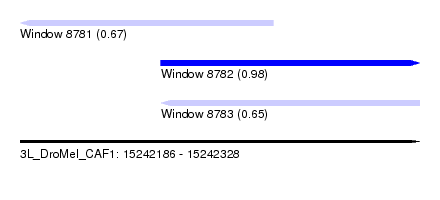
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,242,186 – 15,242,328 |
| Length | 142 |
| Max. P | 0.981423 |

| Location | 15,242,186 – 15,242,276 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 99.11 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -22.11 |
| Energy contribution | -22.11 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15242186 90 - 23771897 GCCGGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUUUGUACAUUAAGUUUUCCCAUCCGCGGCAGUUGACUUUGUUCAGCAUCCUU (((((((((((((....))))))).)).((((((...))))))....................)))).(((((......)))))...... ( -22.40) >DroSec_CAF1 131516 90 - 1 GCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUUUGUACAUUAAGUUUUCCCAUCCGCGGCAGUUGACUUUGUUCAGCAUCCUU (((...(((((((....))))))).((((........(((((.....)))))........))))))).(((((......)))))...... ( -22.09) >DroSim_CAF1 139320 90 - 1 GCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUUUGUACAUUAAGUUUUCCCAUCCGCGGCAGUUGACUUUGUUCAGCAUCCUU (((...(((((((....))))))).((((........(((((.....)))))........))))))).(((((......)))))...... ( -22.09) >DroEre_CAF1 131634 90 - 1 GCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUUUGUACAUUAAGUUUUCCCAUCCGCGGCAGUUGAAUUUGUUCAGCAUCCUU (((...(((((((....))))))).((((........(((((.....)))))........))))))).((((((....))))))...... ( -23.19) >DroYak_CAF1 137434 90 - 1 GCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUUUGUACAUUAAGUUUUCCCAUCCGCGGCAGUUGACUUUGUUCAGCAUCCUU (((...(((((((....))))))).((((........(((((.....)))))........))))))).(((((......)))))...... ( -22.09) >consensus GCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUUUGUACAUUAAGUUUUCCCAUCCGCGGCAGUUGACUUUGUUCAGCAUCCUU (((...(((((((....))))))).((((........(((((.....)))))........))))))).(((((......)))))...... (-22.11 = -22.11 + -0.00)

| Location | 15,242,236 – 15,242,328 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -19.51 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15242236 92 + 23771897 AAUAUAAUUUGCCGCACACACAAACACUUAUGUGGCCGGCACCCCUCGGACCCUCCGAAGGCCCCAAGCCCCAAGCCCCAUUCCUCCAUUCC ..........((((((..............)))))).(((.....((((.....)))).(((.....)))....)))............... ( -19.94) >DroSec_CAF1 131566 85 + 1 AAUAUAAUUUGCCGCACACACAAACACUUAUGUGGCUGGCACCCCUUGGACCCUCCGAAGGCCCCA-------AGCCCCAUUCCCCCAUUCC .........((((((.((((.((....)).)))))).))))...(((((..(((....)))..)))-------))................. ( -19.20) >DroSim_CAF1 139370 85 + 1 AAUAUAAUUUGCCGCACACACAAACACUUAUGUGGCUGGCACCCCUUGGACCCUCCAAAGGCCCCA-------AGUCCCAUUCCCCCAUUCC .........((((((.((((.((....)).)))))).))))...(((((..(((....)))..)))-------))................. ( -19.40) >consensus AAUAUAAUUUGCCGCACACACAAACACUUAUGUGGCUGGCACCCCUUGGACCCUCCGAAGGCCCCA_______AGCCCCAUUCCCCCAUUCC .........((((((.((((.((....)).)))))).)))).((.((((.....)))).))............................... (-17.64 = -17.20 + -0.44)

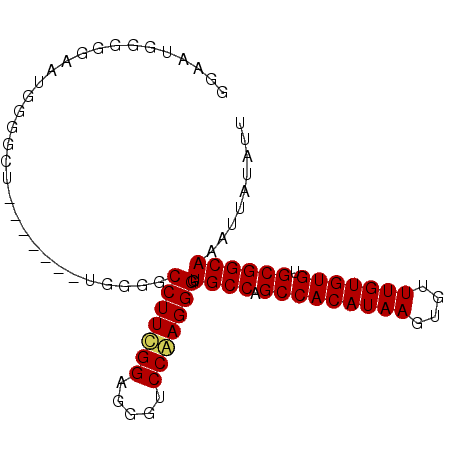
| Location | 15,242,236 – 15,242,328 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15242236 92 - 23771897 GGAAUGGAGGAAUGGGGCUUGGGGCUUGGGGCCUUCGGAGGGUCCGAGGGGUGCCGGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUU (((.((((((..(.((((.....)))).)..)))))).....)))..((..(((((..(((((((....)))))))..)))))..))..... ( -32.20) >DroSec_CAF1 131566 85 - 1 GGAAUGGGGGAAUGGGGCU-------UGGGGCCUUCGGAGGGUCCAAGGGGUGCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUU .................((-------((((.(((....))).))))))...((((.(((((((((....))))))).))))))......... ( -27.40) >DroSim_CAF1 139370 85 - 1 GGAAUGGGGGAAUGGGACU-------UGGGGCCUUUGGAGGGUCCAAGGGGUGCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUU .................((-------((((.(((....))).))))))...((((.(((((((((....))))))).))))))......... ( -27.70) >consensus GGAAUGGGGGAAUGGGGCU_______UGGGGCCUUCGGAGGGUCCAAGGGGUGCCAGCCACAUAAGUGUUUGUGUGUGCGGCAAAUUAUAUU ...............................(((((((.....))))))).((((.(((((((((....))))))).))))))......... (-24.49 = -24.60 + 0.11)


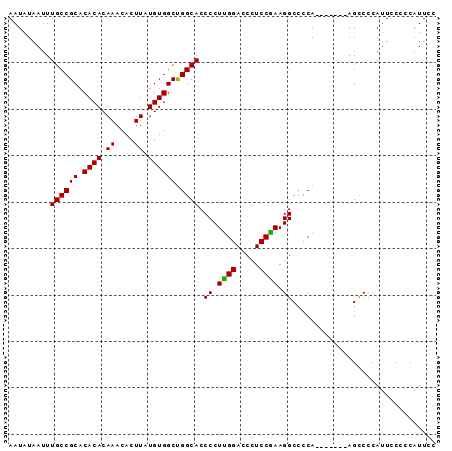
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:13 2006