
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,239,023 – 15,239,124 |
| Length | 101 |
| Max. P | 0.779711 |

| Location | 15,239,023 – 15,239,124 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -10.32 |
| Energy contribution | -10.42 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
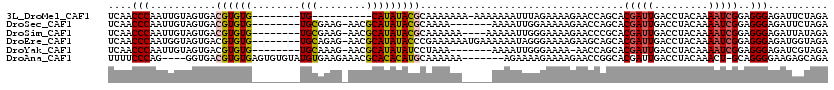
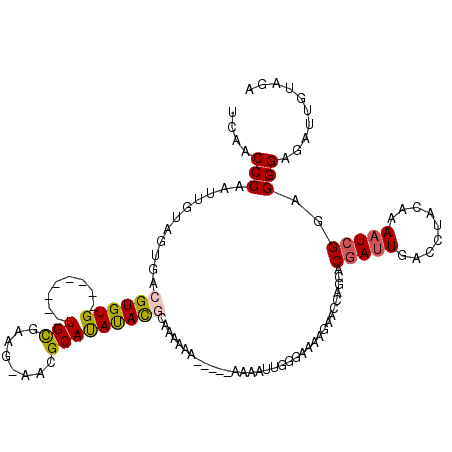
>3L_DroMel_CAF1 15239023 101 + 23771897 UCAACCCAAUUGUAGUGACGUGUG--------UG----------CAUAUACGCAAAAAAA-AAAAAAAUUUAGAAAAGAACCAGCACGAUUGACCUACAAAAUCGGAGGGAGAUUCUAGA ...................(((((--------(.----------...)))))).......-.......(((((((.....((..(.(((((.........))))))..))...))))))) ( -14.80) >DroSec_CAF1 128268 104 + 1 UCAACCCAAUUGUAGUGACGUGUG--------UGCGAAG-AACGCAUAUACGCAAAA-------AAAAUUGGAAAAAGAACCAGCACGAUUGACCUACAAAAUCGGAGGGAGAUUCUAGA .....((((((....((.((((((--------((((...-..))))))))))))...-------..))))))....((((((..(.(((((.........))))))..))...))))... ( -29.60) >DroSim_CAF1 136094 107 + 1 UCAACCCAAUUGUAGUGACGUGUG--------UGCGAAG-AACGCAUAUACGCAAAAAA----AAAAAUUGGGAAAAGAACCCGCACGAUUGACCUACAAAAUCGGAGGGAGAUUAUAGA ....(((((((....((.((((((--------((((...-..)))))))))))).....----...))))))).......(((...(((((.........)))))..))).......... ( -33.70) >DroEre_CAF1 128159 111 + 1 UCAACCCAAUGGUAGUGACGUGUG--------UGCAGAG-AACGCAUAUACCCGAAAAAAUGAAAAAAUAGGGAAAAGAAGCAGCACGAUUGACCUACAAAAUCGGAGGGAGAUGGUAGA ......((((.((.((..(.((((--------(((....-...)))))))(((.................))).......)..)))).))))..((((...(((.......))).)))). ( -21.03) >DroYak_CAF1 134160 103 + 1 UCAACCCAAUUGUAGUGACGUGUG--------UGCAAAG-AACGCAUAUAUCCUAAA-------AAAAUUGGGAAAA-AACCAGCACGAUUGACCUACAAAAUCGGAGGGAGAUCGUAGA ....(((((((.(((.((..((((--------((.....-..))))))..)))))..-------..)))))))....-.......((((((..(((.(......).)))..))))))... ( -30.70) >DroAna_CAF1 121417 108 + 1 UUUUCCCAG----GGUGACGUGUGAGUGUGUAUGUGAAGAAACGCACACAUGCAAAAAA-------AGAAAAGAAAAGAACCGGCACGAUUGACCUACAAACU-GCAGGGGAAGAGCAGA (((((((.(----(((.((((((..((((((.((((......)))).))))))......-------......(........).)))))..).)))).....(.-...))))))))..... ( -25.10) >consensus UCAACCCAAUUGUAGUGACGUGUG________UGCGAAG_AACGCAUAUACGCAAAAAA_____AAAAUUGGGAAAAGAACCAGCACGAUUGACCUACAAAAUCGGAGGGAGAUUGUAGA ....(((...........((((((........(((........)))))))))..................................(((((.........)))))..))).......... (-10.32 = -10.42 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:05 2006