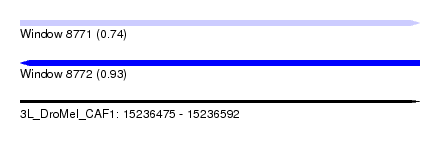
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,236,475 – 15,236,592 |
| Length | 117 |
| Max. P | 0.930276 |

| Location | 15,236,475 – 15,236,592 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
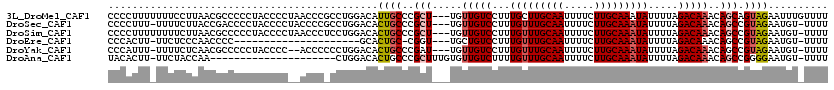
| Mean pairwise identity | 79.92 |
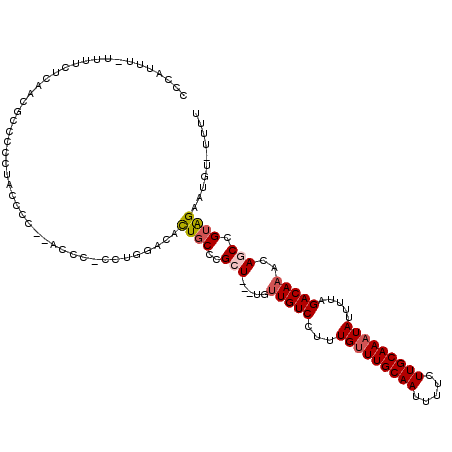
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -15.68 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15236475 117 + 23771897 AAAACAAAUUCUACUGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAGCAAAGGACAACA---AGCGGGCAAUGUCCAGGCGGGUUAGGGGUAGGGGGCGUUAAGGAAAAAAAGGGG ........((((.(((((..((((((......(((((((.....)))))))....))))))..---)))))..(((((((..((.........))...)))))))..))))......... ( -29.00) >DroSec_CAF1 125802 115 + 1 AAAA-ACAUUCUACGGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAACAAAGGACAACA---AGCGGGCAGUGUCCAGGCGGGGUAGGGGUAGGGGUCGGUAAGAAAA-AAAGGGG ....-((.((((((.(((..((((((......(((((((.....)))))))....))))))..---)))((((...)))).......)))))))).................-....... ( -24.70) >DroSim_CAF1 133644 116 + 1 AAAA-ACAUUCUACGGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAACAAAGGACAACA---AGCGGGCAGUGUCCAGGAGGGUUAGGGGUAGGGGGCGUUAAGAAAAAAAAGGGG ...(-((.((((((.(((..((((((......(((((((.....)))))))....))))))..---)))((((...)))).............))))))...)))............... ( -25.00) >DroEre_CAF1 125756 93 + 1 AAAA-ACAUUCUACGGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAACAAAGGACAGCA---ACCG-GCAGUGC---------------------GGGGUUGGGAGAA-AAGUGGG ....-.....((((.(((((((..........(((((((.....)))))))....))))))).---.(((-((.....---------------------...))))).....-..)))). ( -22.24) >DroYak_CAF1 131577 113 + 1 AAAA-ACAUUCUACGGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAACAAAGGACAACA---AUCGGGCAGUGUCCAGGGGGGU--GGGGUAGGGGGCGUUGAGAAAA-AAAUGGG ....-..((((((((..((((.((((......(((((((.....)))))))....))))))))---..)((((...))))......))--)))))......((((.......-.)))).. ( -24.80) >DroAna_CAF1 118803 97 + 1 AAAA-ACAUUCCCCGGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAACAAAAGACAACACAAAGCGGGCAGUGUCCAG---------------------UUGGUAGAA-AAGUGUA ....-(((((.((((..(((((((((......(((((((.....)))))))....)))))).)))...)))).)))))((..---------------------..)).....-....... ( -23.30) >consensus AAAA_ACAUUCUACGGCUGUUUGUCUAAAAUAUUUGCAAGAAAAUUGCAAACAAAGGACAACA___AGCGGGCAGUGUCCAGG_GGGU__GGGGUAGGGGGCGUUAAGAAAA_AAAGGGG ........((((...(((..((((((......(((((((.....)))))))....)))))).....)))((((...))))..........................)))).......... (-15.68 = -15.12 + -0.56)
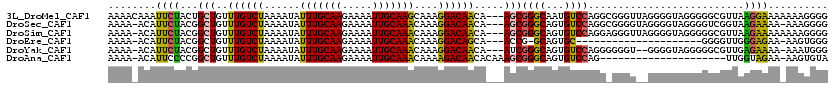
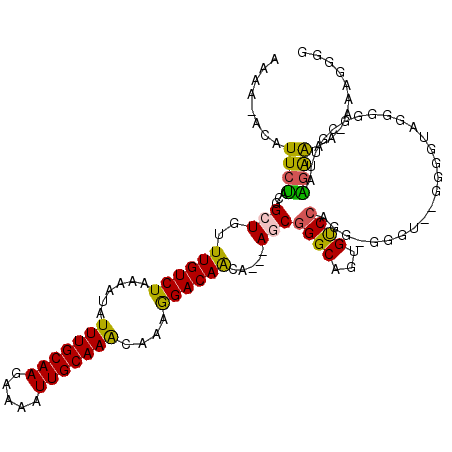
| Location | 15,236,475 – 15,236,592 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.92 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15236475 117 - 23771897 CCCCUUUUUUUCCUUAACGCCCCCUACCCCUAACCCGCCUGGACAUUGCCCGCU---UGUUGUCCUUUGCUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCAGUAGAAUUUGUUUU .......................((((.............((.......))(((---..(((((...((.((((((.....)))))).)).....)))))..))).)))).......... ( -16.70) >DroSec_CAF1 125802 115 - 1 CCCCUUU-UUUUCUUACCGACCCCUACCCCUACCCCGCCUGGACACUGCCCGCU---UGUUGUCCUUUGUUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCCGUAGAAUGU-UUUU .......-................................(((((((((..(((---..(((((...(((((((((.....))))))))).....)))))..))).))))..)))-)).. ( -20.40) >DroSim_CAF1 133644 116 - 1 CCCCUUUUUUUUCUUAACGCCCCCUACCCCUAACCCUCCUGGACACUGCCCGCU---UGUUGUCCUUUGUUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCCGUAGAAUGU-UUUU ..................................((....))...((((..(((---..(((((...(((((((((.....))))))))).....)))))..))).)))).....-.... ( -20.50) >DroEre_CAF1 125756 93 - 1 CCCACUU-UUCUCCCAACCCC---------------------GCACUGC-CGGU---UGCUGUCCUUUGUUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCCGUAGAAUGU-UUUU .......-.............---------------------...((((-.(((---((.((((...(((((((((.....))))))))).....))))..))))))))).....-.... ( -22.00) >DroYak_CAF1 131577 113 - 1 CCCAUUU-UUUUCUCAACGCCCCCUACCCC--ACCCCCCUGGACACUGCCCGAU---UGUUGUCCUUUGUUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCCGUAGAAUGU-UUUU .......-...............((((.((--(......))).........(..---(((((((...(((((((((.....))))))))).....))).))))..))))).....-.... ( -18.90) >DroAna_CAF1 118803 97 - 1 UACACUU-UUCUACCAA---------------------CUGGACACUGCCCGCUUUGUGUUGUCUUUUGUUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCCGGGGAAUGU-UUUU .......-.........---------------------..(((((.(.((((..((((.((((((..(((((((((.....)))))))))....)))))))))).)))).).)))-)).. ( -27.30) >consensus CCCAUUU_UUUUCUCAACGCCCCCUACCCC__ACCC_CCUGGACACUGCCCGCU___UGUUGUCCUUUGUUUGCAAUUUUCUUGCAAAUAUUUUAGACAAACAGCCGUAGAAUGU_UUUU .............................................((((..(((.....(((((...(((((((((.....))))))))).....)))))..))).)))).......... (-13.98 = -14.53 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:03 2006