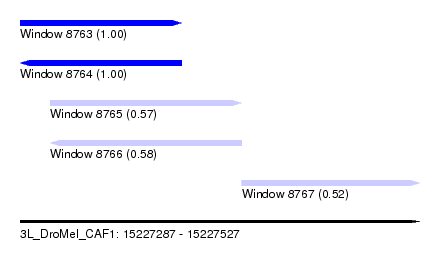
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,227,287 – 15,227,527 |
| Length | 240 |
| Max. P | 0.999315 |

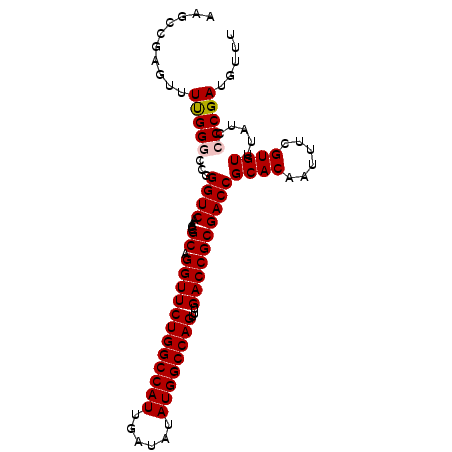
| Location | 15,227,287 – 15,227,384 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -31.24 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
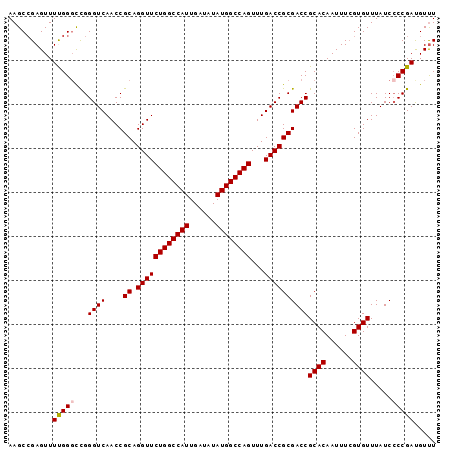
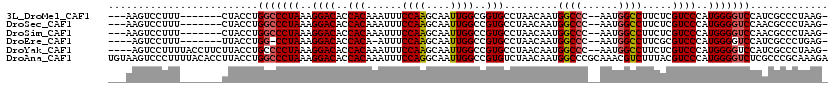
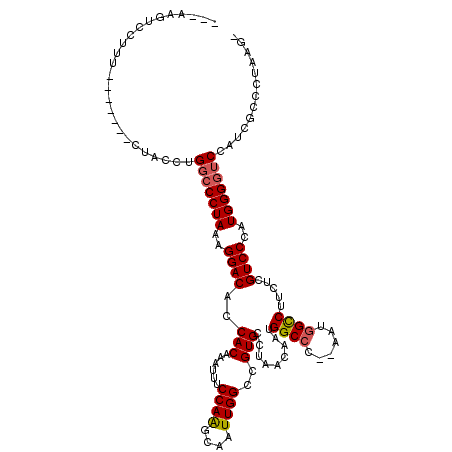
>3L_DroMel_CAF1 15227287 97 + 23771897 AAGCCGAGUUUUGGGCCGGGUCAACCGCAGGUUCUGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUU ..(((........)))((((.....(((.((((((((((((......))))))))...)))))))...((((......)))).....))))...... ( -34.90) >DroSec_CAF1 116565 97 + 1 AAGCCGAGUUUUGGGCCGGGUCAACCGCAGGUUCUGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUGGUGUUUAUCCCCGAUGUUU ..((((((((.((((.((((((((((...))).((((((((......))))))))..))))).)).)).)).))))))))................. ( -35.70) >DroSim_CAF1 124389 97 + 1 AAGCCGAGUUUUGGGCCGGGUCAACCGCAGGUUCUGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUU ..(((........)))((((.....(((.((((((((((((......))))))))...)))))))...((((......)))).....))))...... ( -34.90) >DroEre_CAF1 116758 96 + 1 AAGCGGAGUUUCGGGCUGGGUCAACCGCAGGUUCUGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUC-CCGACGUCU ....(((...(((((...((((....((.((((((((((((......))))))))...))))))))))((((......))))....)-))))..))) ( -34.60) >DroYak_CAF1 122040 97 + 1 AAGCCGAGUUUCGGCUUGGGUCAACCGCAGGUUCUGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUU ((((((.....))))))(((.....(((.((((((((((((......))))))))...)))))))...((((......)))).....)))....... ( -37.90) >consensus AAGCCGAGUUUUGGGCCGGGUCAACCGCAGGUUCUGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUU ..........(((((...((((....((.((((((((((((......))))))))...))))))))))((((......)))).....)))))..... (-31.24 = -31.40 + 0.16)

| Location | 15,227,287 – 15,227,384 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -31.30 |
| Energy contribution | -32.14 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15227287 97 - 23771897 AAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCAGAACCUGCGGUUGACCCGGCCCAAAACUCGGCUU .....(((((......((((....)))).(((((.((((...((((((((......))))))))((((...))))))))))))).....)))))... ( -37.00) >DroSec_CAF1 116565 97 - 1 AAACAUCGGGGAUAAACACCAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCAGAACCUGCGGUUGACCCGGCCCAAAACUCGGCUU .....(((((......(((......))).(((((.((((...((((((((......))))))))((((...))))))))))))).....)))))... ( -36.30) >DroSim_CAF1 124389 97 - 1 AAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCAGAACCUGCGGUUGACCCGGCCCAAAACUCGGCUU .....(((((......((((....)))).(((((.((((...((((((((......))))))))((((...))))))))))))).....)))))... ( -37.00) >DroEre_CAF1 116758 96 - 1 AGACGUCGG-GAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCAGAACCUGCGGUUGACCCAGCCCGAAACUCCGCUU .((..((((-(.....((((....))))((..(((((.....((((((((......))))))))...)))))..))......)))))...))..... ( -33.50) >DroYak_CAF1 122040 97 - 1 AAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCAGAACCUGCGGUUGACCCAAGCCGAAACUCGGCUU .......(((......((((....))))((..(((((.....((((((((......))))))))...)))))..)).)))((((((.....)))))) ( -38.80) >consensus AAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCAGAACCUGCGGUUGACCCGGCCCAAAACUCGGCUU .....(((((......(((......))).(((((.((((...((((((((......))))))))((((...))))))))))))).....)))))... (-31.30 = -32.14 + 0.84)

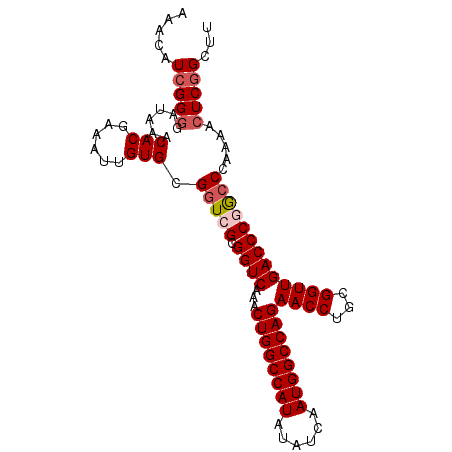
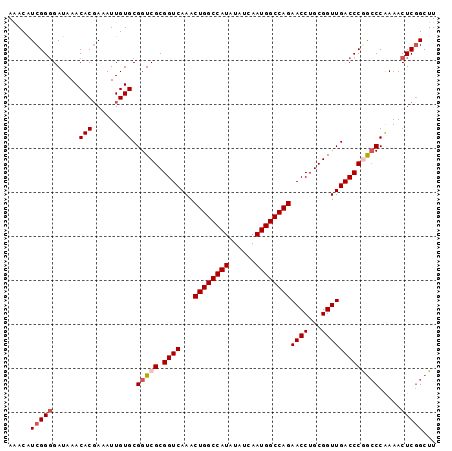
| Location | 15,227,305 – 15,227,420 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15227305 115 + 23771897 GGUCAACCGCAGGUUC-UGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUUGUAAUUUUUUUUGUUCCGCCAUCGAUCUUGGCCGCA---- ((((((.(((.(((((-(((((((......))))))))...)))))))...((((......)))).......(((((..((.(((.......))).)).)))))...))))))...---- ( -35.50) >DroSec_CAF1 116583 115 + 1 GGUCAACCGCAGGUUC-UGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUGGUGUUUAUCCCCGAUGUUUGUAAUUUUUUUUGUUCCGCCAUCGAUCUUGGCCGCA---- ((((....((.(((((-(((((((......))))))))...))))))))))((((((.((((.(.....).))))...))))...............((((.......)))).)).---- ( -36.20) >DroSim_CAF1 124407 115 + 1 GGUCAACCGCAGGUUC-UGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUUGUAAUUUUUUUUGUUCCGCCAUCGAUCUUGGCCGCA---- ((((((.(((.(((((-(((((((......))))))))...)))))))...((((......)))).......(((((..((.(((.......))).)).)))))...))))))...---- ( -35.50) >DroEre_CAF1 116776 114 + 1 GGUCAACCGCAGGUUC-UGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUC-CCGACGUCUAUAAUGUUUUUUGUUCCGCCAGCGAUCUUGGCCGCA---- ((((((.(((.(((((-(((((((......))))))))...))))(((...((((......)))).....-..(((((.....)))))........)))..)))...))))))...---- ( -36.80) >DroYak_CAF1 122058 115 + 1 GGUCAACCGCAGGUUC-UGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUUAUAAUUUUUUUUGUUCCGCCAUCGAUCUUGGCCGCA---- ((((((.(((.(((((-(((((((......))))))))...)))))))...((((......)))).......(((((...((((......)))).....)))))...))))))...---- ( -36.70) >DroAna_CAF1 111136 108 + 1 GGUCAACCGCAGGUUCUUGGCCAUUGAUAUAUGGCCACGUUGACCGCGACCACACAAUCU------------CGACGUUUAUAAUUUUUUUUGUUCCGCCAUCGAUCUUCGCUGAACGCA (((((((..........(((((((......))))))).)))))))(((...........(------------(((.((..((((......))))...))..))))((......)).))). ( -26.50) >consensus GGUCAACCGCAGGUUC_UGGCCAUUGAUAUAUGGCCAGUUUGACCGCGACCGCACAAUUUCGUGUUUAUCCCCGAUGUUUAUAAUUUUUUUUGUUCCGCCAUCGAUCUUGGCCGCA____ ((((((.(((.((((..(((((((......)))))))....)))))))...((((......)))).......(((((...((((......)))).....)))))...))))))....... (-28.16 = -29.10 + 0.94)

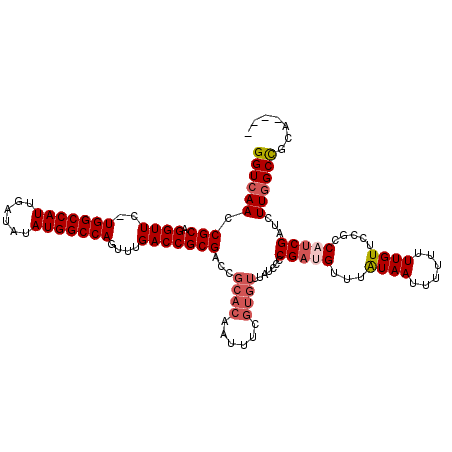

| Location | 15,227,305 – 15,227,420 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -28.82 |
| Energy contribution | -29.93 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15227305 115 - 23771897 ----UGCGGCCAAGAUCGAUGGCGGAACAAAAAAAAUUACAAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCA-GAACCUGCGGUUGACC ----.((((((....((((((......................)))))).......((((....)))).))))))((((...((((((((......)))))))-)((((...)))))))) ( -39.35) >DroSec_CAF1 116583 115 - 1 ----UGCGGCCAAGAUCGAUGGCGGAACAAAAAAAAUUACAAACAUCGGGGAUAAACACCAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCA-GAACCUGCGGUUGACC ----.((((((.....(((((......................)))))((........)).........))))))((((...((((((((......)))))))-)((((...)))))))) ( -39.15) >DroSim_CAF1 124407 115 - 1 ----UGCGGCCAAGAUCGAUGGCGGAACAAAAAAAAUUACAAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCA-GAACCUGCGGUUGACC ----.((((((....((((((......................)))))).......((((....)))).))))))((((...((((((((......)))))))-)((((...)))))))) ( -39.35) >DroEre_CAF1 116776 114 - 1 ----UGCGGCCAAGAUCGCUGGCGGAACAAAAAACAUUAUAGACGUCGG-GAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCA-GAACCUGCGGUUGACC ----.((((((...(((.((((((...................))))))-)))...((((....)))).))))))((((...((((((((......)))))))-)((((...)))))))) ( -42.91) >DroYak_CAF1 122058 115 - 1 ----UGCGGCCAAGAUCGAUGGCGGAACAAAAAAAAUUAUAAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCA-GAACCUGCGGUUGACC ----.((((((....((((((......................)))))).......((((....)))).))))))((((...((((((((......)))))))-)((((...)))))))) ( -39.35) >DroAna_CAF1 111136 108 - 1 UGCGUUCAGCGAAGAUCGAUGGCGGAACAAAAAAAAUUAUAAACGUCG------------AGAUUGUGUGGUCGCGGUCAACGUGGCCAUAUAUCAAUGGCCAAGAACCUGCGGUUGACC .(((..((((((...((((((......................)))))------------)..)))).))..)))(((((((.(((((((......))))))).(......).))))))) ( -33.75) >consensus ____UGCGGCCAAGAUCGAUGGCGGAACAAAAAAAAUUACAAACAUCGGGGAUAAACACGAAAUUGUGCGGUCGCGGUCAAACUGGCCAUAUAUCAAUGGCCA_GAACCUGCGGUUGACC .....((((((....((((((......................)))))).......(((......))).))))))((((....(((((((......)))))))..((((...)))))))) (-28.82 = -29.93 + 1.11)

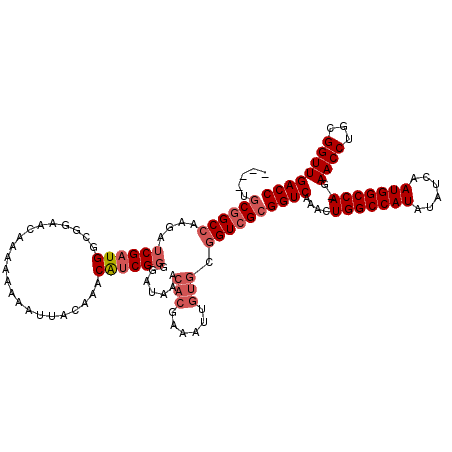
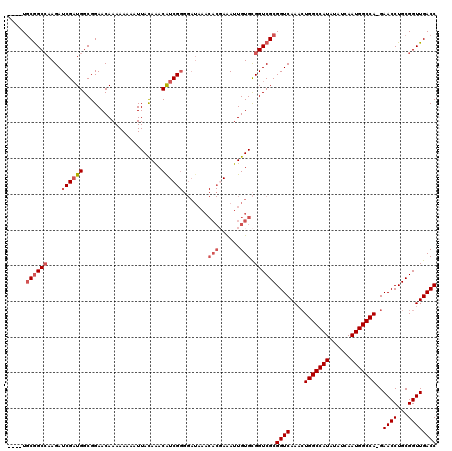
| Location | 15,227,420 – 15,227,527 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -24.21 |
| Energy contribution | -25.68 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15227420 107 + 23771897 ---AAGUCCUUU-------CUACCUGGCCCUAAAGGACACCACAAAUUUCCAAGCAAUUGGCGGUGCCUAACAAUGGCCC--AAUGGCCUUCUCGUCCCAUGGGGUCCAUCGCCCUAAG- ---.........-------.....((((((((..((((.......((..((((....))))..))..........((((.--...)))).....))))..))))).)))..........- ( -29.80) >DroSec_CAF1 116698 107 + 1 ---AAGUCCUUU-------CUACCUGGCCCUAAAGGACACCACAAAUUUCCAAGCAAUUGGCCGUGCCUAACAAUGGCCC--AAUGGCCUUCUCGUCCCAUGGGGUCCAACGCCCUAAG- ---.........-------.....((((((((..((((.............((....))(((((((((.......)))..--.)))))).....))))..))))).)))..........- ( -30.00) >DroSim_CAF1 124522 107 + 1 ---AAGUCCUUU-------CUACCUGGCCCUAAAGGACACCACAAAUUUCCAAGCAAUUGGCCGUGCCUAACAAUGGCCC--AAUGGCCUUCUCGUCCCAUGGGGUCCAACGCCCUAAG- ---.........-------.....((((((((..((((.............((....))(((((((((.......)))..--.)))))).....))))..))))).)))..........- ( -30.00) >DroEre_CAF1 116890 104 + 1 ----AGUCCUUU-------UUACCUGG-CCUAAAGGACACCACA-AUUUCCAAGCAAUUGGCCGUGCCUAACAAUGGCCC--AAUGGCCUUCGCGUCCCAUGGGGUCCAUCGCCCUGAG- ----.(((((((-------........-...)))))))......-........((....(((((((((.......)))..--.))))))...)).......(((((.....)))))...- ( -29.20) >DroYak_CAF1 122173 113 + 1 ----AGUCCUUUUACCUUCUUACCUGCCCCUAAAGGACACCACAAAUUUCCAAGCAAUUGGCCGUGCCUAACAAUGGCCC--AAUGGCCUUCUCGUCCCAUGGGGUCCAUCGCCCUAAG- ----.(((((((...................))))))).....................(((((((((.......)))..--.))))))...........((((((.....))))))..- ( -27.71) >DroAna_CAF1 111244 120 + 1 UGUAAGUCCCUUUUACACCUUACCUGGCCCUAAAGGACACCACAAAUUUCCAGGCAAUUGGCCGUGUCUAACAAUGGCCCGCAAACGUCUUUACGUCCCAUGGGGUCUCGCCCGCAAAGA ((((((.....))))))........(((((((..((((...............((....((((((........)))))).))....((....))))))..)))))))............. ( -29.90) >consensus ___AAGUCCUUU_______CUACCUGGCCCUAAAGGACACCACAAAUUUCCAAGCAAUUGGCCGUGCCUAACAAUGGCCC__AAUGGCCUUCUCGUCCCAUGGGGUCCAUCGCCCUAAG_ .........................(((((((..((((..(((......((((....))))..))).........((((......)))).....))))..)))))))............. (-24.21 = -25.68 + 1.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:58 2006