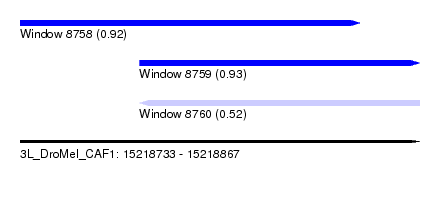
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,218,733 – 15,218,867 |
| Length | 134 |
| Max. P | 0.934859 |

| Location | 15,218,733 – 15,218,847 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.16 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -25.26 |
| Energy contribution | -26.98 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15218733 114 + 23771897 AACUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCCGG-- ..((((((((....))))(((((((....))).))))))))...((.((((..((((((((....(((((....)))))....(((.......))).---))))))))..)))))).-- ( -30.70) >DroSec_CAF1 108259 114 + 1 AACUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG-- ..((((((((....))))(((((((....))).))))))))......((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...-- ( -30.70) >DroSim_CAF1 115584 114 + 1 AAAUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG-- .(((((.......)))))(((((((....))).))))..........((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...-- ( -30.40) >DroEre_CAF1 108493 114 + 1 AACUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG-- ..((((((((....))))(((((((....))).))))))))......((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...-- ( -30.70) >DroYak_CAF1 112931 114 + 1 AACUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG-- ..((((((((....))))(((((((....))).))))))))......((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...-- ( -30.70) >DroAna_CAF1 103305 119 + 1 AACUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGUGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGCACCGAGAGAUCCGAGUUAGAAA ((((.(((......((((...((.(((((..((......))..))))))).))))(((((.....(((((....)))))..(((((.......)))))..))))).))).))))..... ( -27.00) >consensus AACUAGGACAAAACUGUUUUUGGUCACAUGACUCAAAUUAGAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA___GGGAAAUGAGAGGCUGG__ .............(((..(((((((....))).)))).)))......((((..((((((((....(((((....)))))....(((.......)))....))))))))..))))..... (-25.26 = -26.98 + 1.72)


| Location | 15,218,773 – 15,218,867 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -19.06 |
| Energy contribution | -20.78 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
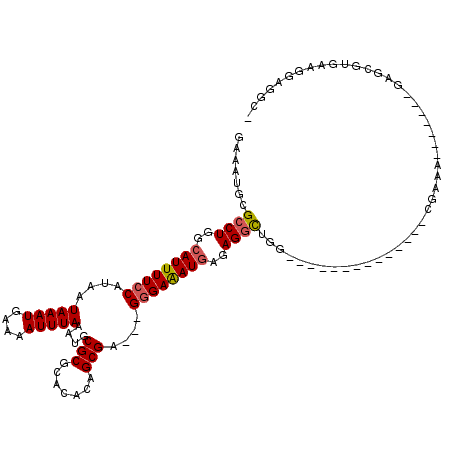
>3L_DroMel_CAF1 15218773 94 + 23771897 GAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCCGG--------------CGAAA-------GAGCGUGAAGGAGGC- ...((((((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...--------------(....-------).)))).........- ( -28.60) >DroSec_CAF1 108299 94 + 1 GAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG--------------CGAAA-------GAGCGUGAAGGAGGC- ...((((((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...--------------(....-------).)))).........- ( -29.00) >DroSim_CAF1 115624 94 + 1 GAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG--------------CGAAA-------GAGCGUGAAGGAGGC- ...((((((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...--------------(....-------).)))).........- ( -29.00) >DroEre_CAF1 108533 80 + 1 GAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG--------------CCAAA-----------------G----- .......((((..((((((((....(((((....)))))....(((.......))).---))))))))..))))...--------------.....-----------------.----- ( -24.10) >DroYak_CAF1 112971 101 + 1 GAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA---GGGAAAUGAGAGGCUGG--------------CUGCAAAUGCGAGAGCGUGAAGGAGGC- .......((((..((((((((....(((((....)))))....(((.......))).---))))))))..)))).(.--------------((.(..((((....))))...).)).)- ( -32.20) >DroAna_CAF1 103345 112 + 1 GAAAUGUGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGCACCGAGAGAUCCGAGUUAGAAAAAAAAAUAAACACAGAA-------GACACUAAAGGAGGUG .......(((.....(((((.....(((((....)))))..(((((.......)))))..))))).(((...((((....................-------....)))).)))))). ( -18.06) >consensus GAAAUGCGCCUGGCAUUUUCCAUAAUAAAUGAAAAUUUAAAUGCGCGCACACAGCGA___GGGAAAUGAGAGGCUGG______________CGAAA_______GAGCGUGAAGGAGGC_ .......((((..((((((((....(((((....)))))....(((.......)))....))))))))..))))............................................. (-19.06 = -20.78 + 1.72)


| Location | 15,218,773 – 15,218,867 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
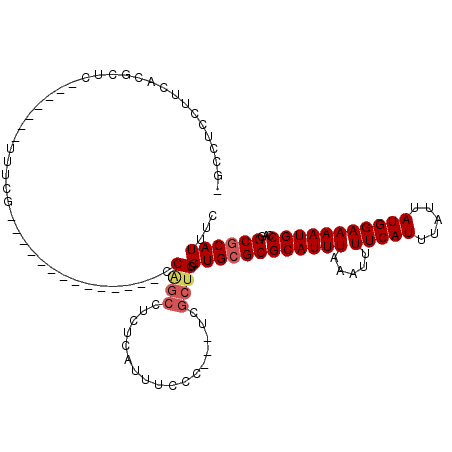
>3L_DroMel_CAF1 15218773 94 - 23771897 -GCCUCCUUCACGCUC-------UUUCG--------------CCGGCCUCUCAUUUCCC---UCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCGCAUUUC -((((.......((..-------....)--------------).(((...........(---.(((.....))).)........(((((((.....)))))))..)))))))....... ( -17.70) >DroSec_CAF1 108299 94 - 1 -GCCUCCUUCACGCUC-------UUUCG--------------CCAGCCUCUCAUUUCCC---UCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCGCAUUUC -((((.......((..-------...((--------------(((((............---..)))).))).))((((((.....(((((.....))))))))))).))))....... ( -17.94) >DroSim_CAF1 115624 94 - 1 -GCCUCCUUCACGCUC-------UUUCG--------------CCAGCCUCUCAUUUCCC---UCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCGCAUUUC -((((.......((..-------...((--------------(((((............---..)))).))).))((((((.....(((((.....))))))))))).))))....... ( -17.94) >DroEre_CAF1 108533 80 - 1 -----C-----------------UUUGG--------------CCAGCCUCUCAUUUCCC---UCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCGCAUUUC -----.-----------------....(--------------(((((............---..)))).))((((((((((.....(((((.....)))))))))))...))))..... ( -16.54) >DroYak_CAF1 112971 101 - 1 -GCCUCCUUCACGCUCUCGCAUUUGCAG--------------CCAGCCUCUCAUUUCCC---UCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCGCAUUUC -((((.......((...(((((..((((--------------(.((............)---).)))))))))).)).......(((((((.....))))))).....))))....... ( -22.70) >DroAna_CAF1 103345 112 - 1 CACCUCCUUUAGUGUC-------UUCUGUGUUUAUUUUUUUUUCUAACUCGGAUCUCUCGGUGCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCACAUUUC ...........(((((-------(((((.(((.............))).))))......(((((((.....)))).........(((((((.....)))))))..)))))))))..... ( -25.72) >consensus _GCCUCCUUCACGCUC_______UUUCG______________CCAGCCUCUCAUUUCCC___UCGCUGUGUGCGCGCAUUUAAAUUUUCAUUUAUUAUGGAAAAUGCCAGGCGCAUUUC ...........................................((((.................)))).((((((((((((.....(((((.....)))))))))))...))))))... (-13.14 = -13.70 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:52 2006