| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,213,908 – 15,214,032 |
| Length | 124 |
| Max. P | 0.587402 |

| Location | 15,213,908 – 15,213,998 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -14.42 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.45 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15213908 90 + 23771897 UUUAUUUUCCA-------CCGAUUCU---CUUCUGUGGCUUU-UUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUA ........(((-------(.((....---..)).))))....-.....(((((((((((((((((......))))))).))))..)))))).......... ( -17.10) >DroVir_CAF1 93032 79 + 1 -------GUUG-------CUGUUGUUGUU-----GUUUU--U-UUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUA -------...(-------((...((((.(-----((...--.-...))).))))(((((((((((......))))))).)))).....))).......... ( -12.90) >DroSec_CAF1 103415 79 + 1 ------UUUCA-------CCGAUU--------CUGUGGCUUU-UUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUA ------...((-------(.....--------..)))((...-...))(((((((((((((((((......))))))).))))..)))))).......... ( -13.60) >DroSim_CAF1 110779 77 + 1 ------UUCCA-------CAAAUU--------CUGAGGCUU--UUUGCGUCAAAGUUAUAAUAGAUCGCAUUCAGUUAUAAACAAGUUGGCUGUUGAU-UA ------.....-------..((((--------.(((.((..--...)).))).))))......(((((....((((((.........)))))).))))-). ( -13.30) >DroYak_CAF1 108081 85 + 1 -----UUUCCA-------CCGAUUCU---CUGCUGUGGCUUU-UUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUA -----...(((-------(.(.....---...).))))....-.....(((((((((((((((((......))))))).))))..)))))).......... ( -14.70) >DroAna_CAF1 98266 92 + 1 ------UUUCUCCGAUUUCUGAUUCU---CGGCUGUGGCUUUUUUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUA ------.....((((..........)---))).....((.......))(((((((((((((((((......))))))).))))..)))))).......... ( -14.90) >consensus ______UUCCA_______CCGAUUCU______CUGUGGCUUU_UUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUA ................................................(((((((((((((((((......))))))).))))..)))))).......... (-10.09 = -10.45 + 0.36)


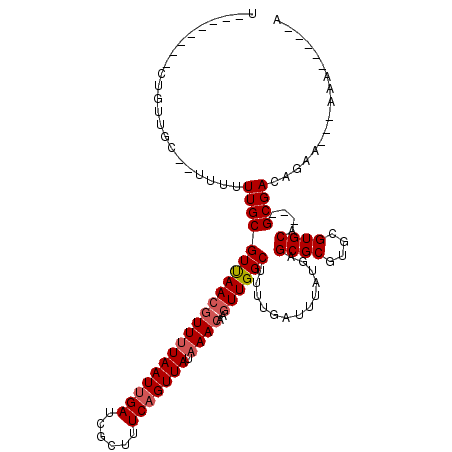
| Location | 15,213,926 – 15,214,032 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15213926 106 + 23771897 U-----CUUCUGUGGC--UUUUUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUAUGAGCGCGUGCGUGCAGCAGCGACAGAGAA-AAA-----A .-----.((((((.((--(.....(((((((((((((((((((......))))))).))))..)))))).............((((....)))).))))).))))))..-...-----. ( -29.40) >DroVir_CAF1 93043 96 + 1 UGUU-------GUUUU----UUUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUAUGAGCGCGUGCGUGCA---GCGACAGAA----AA-----A ....-------.((((----(.(((((((((((((((((((((......))))))).))))..)))))).............((((....)))).---)))).))))----).-----. ( -22.80) >DroPse_CAF1 91529 104 + 1 --------UCUUU--A--UUUUUUGCGUUAACAUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUAUGAGCGCGUGCGUGCA---GCGACAGAAAAAAAAAUUCAA --------.....--.--(((((((((((..(((...(((..(..(((..(............)..)))..)..))).))).((((....)))))---))).))))))).......... ( -19.90) >DroGri_CAF1 99373 108 + 1 UGUUUUAUUUUGUUGCUUUUUUUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUAUGAGCGCGUGCGUGCA---GCGACAGAA---AAA-----A .......((((((((((.........(((((((((((((((((......))))))).))))..)))))).............((((....)))))---)))))))))---...-----. ( -28.10) >DroSim_CAF1 110790 100 + 1 ---------CUGAGGC--UU-UUUGCGUCAAAGUUAUAAUAGAUCGCAUUCAGUUAUAAACAAGUUGGCUGUUGAU-UAUGAGCGCGAGCGUGCAGCCGCGACAGAGAA-AAA-----A ---------.(((.((--..-...)).)))...(((((((.((......)).))))))).........((((((..-..((.((....))...))....))))))....-...-----. ( -23.80) >DroMoj_CAF1 97352 100 + 1 UGUU-------GUUUUUAUUUUUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUAUGAGCGCGUGCGUGCA---GCGACAGAA----AA-----A ((((-------(((............(((((((((((((((((......))))))).))))..)))))).............((((....)))))---))))))...----..-----. ( -22.60) >consensus U________CUGUUGC__UUUUUUGCGUUAACGUUUUAAUUGAUCGCUUUCAGUUAUAAACAAGUUGGCUUUUGAUUUAUGAGCGCGUGCGUGCA___GCGACAGAA___AAA_____A ......................(((((((((((((((((((((......))))))).))))..)))))).............((((....))))....))))................. (-18.56 = -19.08 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:48 2006