
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,212,414 – 15,212,574 |
| Length | 160 |
| Max. P | 0.989573 |

| Location | 15,212,414 – 15,212,534 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15212414 120 + 23771897 AAGAUUAAAUUUAUUAAACUGAUUUUGAAACAACGAAGACAAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAAUGCUUGUUGACGCAGCAAUAAAUUUAAAAUUUUUU ....(((((((((((...(((.(((((......))))).........(((.((((......)))).)))..((((((((((....))))))))))..))).)))))))))))........ ( -21.90) >DroSec_CAF1 101802 118 + 1 AAGAUUAAAUUUAUUAAACUGAUUUCGAAACAACCAAGACAAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAUUGCUUGUUGACGCAGCAAUAAAUUUAAA--UUUUU ....(((((((((((...(((..........................(((.((((......)))).)))..((((((((((....))))))))))..))).))))))))))).--..... ( -21.90) >DroSim_CAF1 107317 118 + 1 AAGAUUAAAUUUAUUAAACUGAUUCCGAAACAACCAAGACAAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAAUGCUUGUUGACGCAGCAAUAAAUUUUAA--UUUUU (((((((((((((((...(((..........................(((.((((......)))).)))..((((((((((....))))))))))..))).)))))).)))))--)))). ( -20.90) >DroEre_CAF1 102293 117 + 1 AAGAUUAAGUUUAUUAAACUGAUUUCCCGACAA-CAAGACAAACCAUAUUUAUGCAAAACAGCGUCAAUCUUUAACAAGCAUGAAUGCUUGUUGACGCAGCUAUAAAUCUCAG--UAUUU .................(((((...........-..........(((....))).......((((((.......(((((((....)))))))))))))...........))))--).... ( -20.01) >DroYak_CAF1 106513 118 + 1 AAGAUUAAAUUUAUUAAACUGAUUUCCAAACAACCAAGACAAACCAUAUUUAUGCAAAGCAGCGCCAAUCUUUAACAAGCAUGAAUGCUUGUUGACGCAGCUAUAAAUCUUCA--UUUUU ((((((......................................(((....)))...(((.(((.......((((((((((....))))))))))))).)))...))))))..--..... ( -18.01) >consensus AAGAUUAAAUUUAUUAAACUGAUUUCGAAACAACCAAGACAAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAAUGCUUGUUGACGCAGCAAUAAAUUUAAA__UUUUU ......(((((((((...(((..........................(((.((((......)))).)))..((((((((((....))))))))))..))).))))))))).......... (-17.00 = -17.60 + 0.60)

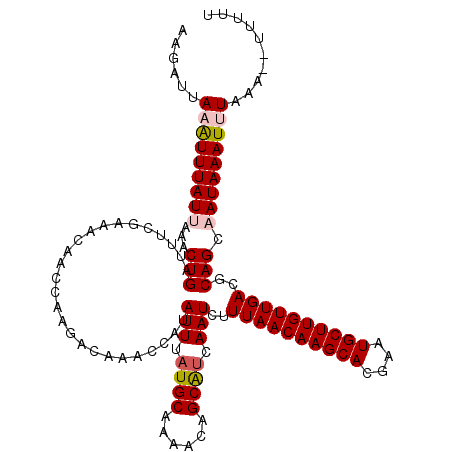

| Location | 15,212,414 – 15,212,534 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15212414 120 - 23771897 AAAAAAUUUUAAAUUUAUUGCUGCGUCAACAAGCAUUCGUGCUUGUUAAAGAUUGAUGCUGUUUUGCAUAAAUAUGGUUUGUCUUCGUUGUUUCAAAAUCAGUUUAAUAAAUUUAAUCUU ........(((((((((((((((((((((((((((....))))))).......)))))).((((((....(((..((.....))..)))....)))))).)))..))))))))))).... ( -23.81) >DroSec_CAF1 101802 118 - 1 AAAAA--UUUAAAUUUAUUGCUGCGUCAACAAGCAAUCGUGCUUGUUAAAGAUUGAUGCUGUUUUGCAUAAAUAUGGUUUGUCUUGGUUGUUUCGAAAUCAGUUUAAUAAAUUUAAUCUU .....--.(((((((((((((((..((((((((((....)))))))).(((((.(((..(((((.....)))))..))).))))).........))...))))..))))))))))).... ( -25.50) >DroSim_CAF1 107317 118 - 1 AAAAA--UUAAAAUUUAUUGCUGCGUCAACAAGCAUUCGUGCUUGUUAAAGAUUGAUGCUGUUUUGCAUAAAUAUGGUUUGUCUUGGUUGUUUCGGAAUCAGUUUAAUAAAUUUAAUCUU .....--...(((((((((((((..((((((((((....)))))))).(((((.(((..(((((.....)))))..))).)))))..........))..))))..)))))))))...... ( -23.70) >DroEre_CAF1 102293 117 - 1 AAAUA--CUGAGAUUUAUAGCUGCGUCAACAAGCAUUCAUGCUUGUUAAAGAUUGACGCUGUUUUGCAUAAAUAUGGUUUGUCUUG-UUGUCGGGAAAUCAGUUUAAUAAACUUAAUCUU .....--.((((.((((((((.(((((((((((((....))))))).......)))))).))).....(((((.((((((.((...-.....)).))))))))))))))))))))..... ( -26.61) >DroYak_CAF1 106513 118 - 1 AAAAA--UGAAGAUUUAUAGCUGCGUCAACAAGCAUUCAUGCUUGUUAAAGAUUGGCGCUGCUUUGCAUAAAUAUGGUUUGUCUUGGUUGUUUGGAAAUCAGUUUAAUAAAUUUAAUCUU .....--...(((((((((((.(((((((((((((....))))))).......)))))).))).....(((((.((((((.((..........)))))))))))))))))))))...... ( -25.51) >consensus AAAAA__UUAAAAUUUAUUGCUGCGUCAACAAGCAUUCGUGCUUGUUAAAGAUUGAUGCUGUUUUGCAUAAAUAUGGUUUGUCUUGGUUGUUUCGAAAUCAGUUUAAUAAAUUUAAUCUU ..........((((((((.((.(((((((((((((....))))))).......)))))).))......(((((.((((((...............)))))))))))))))))))...... (-21.79 = -21.03 + -0.76)


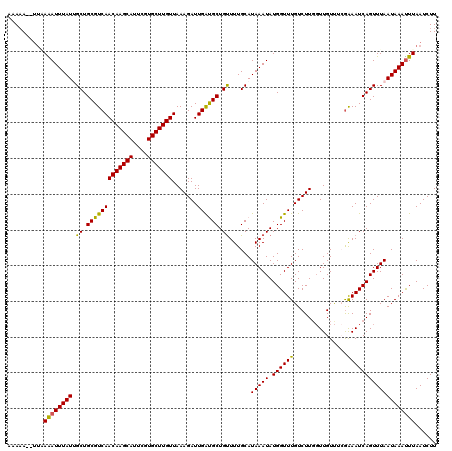
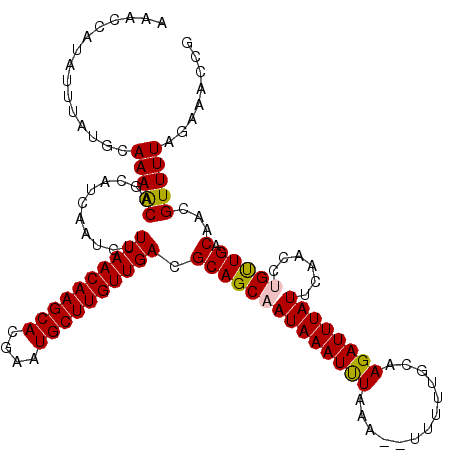
| Location | 15,212,454 – 15,212,574 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -19.18 |
| Energy contribution | -18.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15212454 120 + 23771897 AAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAAUGCUUGUUGACGCAGCAAUAAAUUUAAAAUUUUUUGCAAGAUUUAUUCAACGUGUUGACAAUGUUUUAGAAACCG ...........((((......))))...(((..((((.(((....)))((((..((((.((((.((((.....)))).))))..((.....))...))))..))))))))..)))..... ( -25.10) >DroSec_CAF1 101842 118 + 1 AAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAUUGCUUGUUGACGCAGCAAUAAAUUUAAA--UUUUUGCAAGAUUUAUUCAACCUGUUGACAACGUUUUAAAAACCA ...............(((((......(((((((((((((((....)))))))))).((((.(((........)--)).)))).)))))...((((....))))....)))))........ ( -20.10) >DroSim_CAF1 107357 118 + 1 AAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAAUGCUUGUUGACGCAGCAAUAAAUUUUAA--UUUUUGCAAGAUUUAUUCAACCUGUUGACAACGUUUUAAAAACCG ...............(((((......(((((((((((((((....)))))))))).((((.(((........)--)).)))).)))))...((((....))))....)))))........ ( -20.00) >DroEre_CAF1 102332 118 + 1 AAACCAUAUUUAUGCAAAACAGCGUCAAUCUUUAACAAGCAUGAAUGCUUGUUGACGCAGCUAUAAAUCUCAG--UAUUUGCAAGAUUUAUUCCACCUGCUGACAACGUUUUAGAAACCG ...............(((((...((((....((((((((((....)))))))))).((((..((((((((..(--(....)).)))))))).....))))))))...)))))........ ( -26.50) >DroYak_CAF1 106553 118 + 1 AAACCAUAUUUAUGCAAAGCAGCGCCAAUCUUUAACAAGCAUGAAUGCUUGUUGACGCAGCUAUAAAUCUUCA--UUUUUGCCAGAUUUAUUCAACCUGCUGACAACGUUUUAGAAACCG ...............(((((...........((((((((((....)))))))))).((((..((((((((.((--....))..)))))))).....)))).......)))))........ ( -23.50) >consensus AAACCAUAUUUAUGCAAAACAGCAUCAAUCUUUAACAAGCACGAAUGCUUGUUGACGCAGCAAUAAAUUUAAA__UUUUUGCAAGAUUUAUUCAACCUGUUGACAACGUUUUAGAAACCG ...............(((((...........((((((((((....)))))))))).((((((((((((((.............))))))))......))))).)...)))))........ (-19.18 = -18.94 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:46 2006