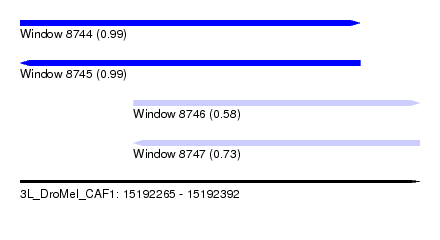
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,192,265 – 15,192,392 |
| Length | 127 |
| Max. P | 0.992845 |

| Location | 15,192,265 – 15,192,373 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15192265 108 + 23771897 ----UGUUGCCCCAUGCCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAACGUCGUCGCCUUUUUUGGCCCAGGCGUCUAAGCC----GG----GACUUGUAAAAGGAUCCUCGGCUC ----.........((((((.((((......))))((((((((..((((((......))))))..)))))))).))))))...((((----((----(((((.....))).))).))))). ( -45.80) >DroVir_CAF1 71150 91 + 1 ----UGUUGC-CCAUGGCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAACGUCGUCGCCUUUUUAGGCCCAGGCGUCUAAGUU----GCAGGCAACU-------------------- ----.(((((-(..(((((((((((......)).((((.(((..((((((......))))))..))).)))).....))).)))))----)..)))))).-------------------- ( -36.80) >DroPse_CAF1 70841 110 + 1 ----UGUUGCCCCAUGCCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAACGUCGUCGCCUUUUUUGGUCCAGGCGUCUAAGCCAGCAGA----CUCUUAUAAAAGGAUCCACU--CU ----............((((.....(((((((((((((((((..((((((......))))))..))))))))..(((......)))))))))----.))).....)))).......--.. ( -37.30) >DroGri_CAF1 77502 99 + 1 UUGCUGUUGC-CCAUGGCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAGCGUCGUCGCCUUUUUAGGCCCAGGCGUCUAAGUU----GCAGGCAACU----------------GCUU ..((((((((-(..(((((.((((......))))))))).....)))))))))(.((((((((....))))...)))).)......----((((....))----------------)).. ( -41.20) >DroMoj_CAF1 76009 91 + 1 ----UGUUGC-CCAUGGCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAACGUCGUCGCCUUUUUAGGCCCAGGCGUCUAUUUC----GCAGCCAACU-------------------- ----......-...((((((((.(.((((.(.(.((((.(((..((((((......))))))..))).)))).).).)))).).))----).)))))...-------------------- ( -33.00) >DroPer_CAF1 70982 110 + 1 ----UGUUGCCCCAUGCCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAACGUCGUCGCCUUUUUUGGUCCAGGCGUCUAAGCCAGCCGA----GUCUUAUAAAAGGAUCCACU--CU ----............((((.....(((((((.(((((((((..((((((......))))))..)))))).)))(((......)))...)))----)))).....)))).......--.. ( -38.70) >consensus ____UGUUGC_CCAUGCCUUGACACAGAUUUGUUGGCCAAAGUUGGCGACAACGUCGUCGCCUUUUUAGGCCCAGGCGUCUAAGCC____GC____AACU___________________U .........................((((.(.(.((((.(((..((((((......))))))..))).)))).).).))))....................................... (-23.42 = -23.20 + -0.22)


| Location | 15,192,265 – 15,192,373 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -20.77 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15192265 108 - 23771897 GAGCCGAGGAUCCUUUUACAAGUC----CC----GGCUUAGACGCCUGGGCCAAAAAAGGCGACGACGUUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGGCAUGGGGCAACA---- ((((((.((((..........)))----))----)))))....(((..(((((((...(((((((....)))))))...))))))).......((((((....)))))))))....---- ( -46.30) >DroVir_CAF1 71150 91 - 1 --------------------AGUUGCCUGC----AACUUAGACGCCUGGGCCUAAAAAGGCGACGACGUUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGCCAUGG-GCAACA---- --------------------.((((((((.----..(((.(((((...((((......(((((((....)))))))......)))).........))))))))...)))-))))).---- ( -35.90) >DroPse_CAF1 70841 110 - 1 AG--AGUGGAUCCUUUUAUAAGAG----UCUGCUGGCUUAGACGCCUGGACCAAAAAAGGCGACGACGUUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGGCAUGGGGCAACA---- ..--((..((..(((....)))..----))..)).........((((((.(((((...(((((((....)))))))...))))))))......((((((....)))))))))....---- ( -38.60) >DroGri_CAF1 77502 99 - 1 AAGC----------------AGUUGCCUGC----AACUUAGACGCCUGGGCCUAAAAAGGCGACGACGCUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGCCAUGG-GCAACAGCAA ..((----------------.((((((((.----..(((.(((((...((((......(((((((....)))))))......)))).........))))))))...)))-))))).)).. ( -38.40) >DroMoj_CAF1 76009 91 - 1 --------------------AGUUGGCUGC----GAAAUAGACGCCUGGGCCUAAAAAGGCGACGACGUUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGCCAUGG-GCAACA---- --------------------...(((((..----((.(((((......((((......(((((((....)))))))......))))......))))).)).)))))..(-....).---- ( -33.90) >DroPer_CAF1 70982 110 - 1 AG--AGUGGAUCCUUUUAUAAGAC----UCGGCUGGCUUAGACGCCUGGACCAAAAAAGGCGACGACGUUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGGCAUGGGGCAACA---- ..--(((.((..(((....)))..----)).))).........((((((.(((((...(((((((....)))))))...))))))))......((((((....)))))))))....---- ( -35.60) >consensus A___________________AGUU____GC____GACUUAGACGCCUGGGCCAAAAAAGGCGACGACGUUGUCGCCAACUUUGGCCAACAAAUCUGUGUCAAGCCAUGG_GCAACA____ ..................................(((.((((......((((......(((((((....)))))))......))))......)))).))).................... (-20.77 = -21.35 + 0.58)



| Location | 15,192,301 – 15,192,392 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -21.95 |
| Energy contribution | -23.82 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15192301 91 + 23771897 AGUUGGCGACAACGUCGUCGCCUUUUUUGGCCCAGGCGUCUAAGCC----GGGACUUGUAAAAGGAUCCUCGGCUCGGCUCGGCUCCUGCGCCUU ....((((((......))))))......(((.((((.(((..((((----(((((((.....))).))).)))))..)....)).)))).))).. ( -38.70) >DroPse_CAF1 70877 89 + 1 AGUUGGCGACAACGUCGUCGCCUUUUUUGGUCCAGGCGUCUAAGCCAGCAGACUCUUAUAAAAGGAUCCACU--CUGGCCUGGCUCCUUCU---- .((((((....(((((...(((......)))...)))))....))))))............(((((.(((..--......))).)))))..---- ( -28.80) >DroSec_CAF1 81835 91 + 1 AGUUGGCGACAACGUCGUCGCCUUUUUUGGCCCAGGCGUCUAAGCC----GGGACUUGUAAAAGGAUCCUCGGCUUGGUUCGGCUCCUGCGCCUU ....((((((......))))))......(((.((((.(((((((((----(((((((.....))).))).))))))))....)).)))).))).. ( -41.60) >DroEre_CAF1 81832 84 + 1 AGUUGGCGACAACGCCGUCGCCUUUUUUGGCCCAGGCGUCUAAGCC----GGGCCUUGUAAAAGGAUGCUCGGC------CGGCUCCUGCGCCU- .((((....))))((((.((((((((..(((((.(((......)))----)))))....)))))).))..))))------.(((......))).- ( -39.80) >DroYak_CAF1 85139 90 + 1 AGUUGGCGACAACGUCGUCGCCUUUUUUGGCCCAGGCGUCUUAGCC----GGGACUUGUAAAAGGAUCCUCGGCUCGGAUCGGCUCCUGCGCCU- ....((((((......))))))......(((.(((((((((.((((----(((((((.....))).))).))))).))).))...)))).))).- ( -39.60) >DroPer_CAF1 71018 89 + 1 AGUUGGCGACAACGUCGUCGCCUUUUUUGGUCCAGGCGUCUAAGCCAGCCGAGUCUUAUAAAAGGAUCCACU--CUGGCCUGGCUCCUUCU---- ....((((((......))))))...((((((...(((......))).))))))........(((((.(((..--......))).)))))..---- ( -28.00) >consensus AGUUGGCGACAACGUCGUCGCCUUUUUUGGCCCAGGCGUCUAAGCC____GGGACUUGUAAAAGGAUCCUCGGCUUGGCUCGGCUCCUGCGCCU_ ....((((((......))))))......(((.((((.(((...(((....((..(((....)))..))........)))..))).)))).))).. (-21.95 = -23.82 + 1.86)

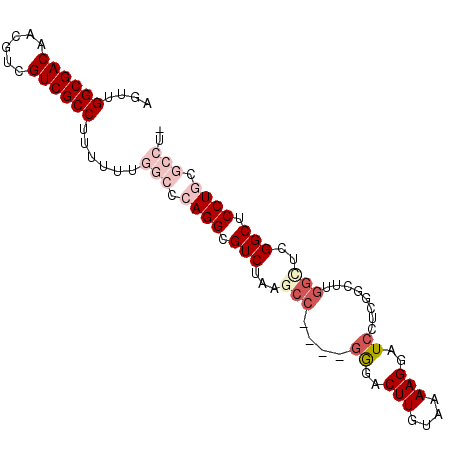
| Location | 15,192,301 – 15,192,392 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -23.52 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15192301 91 - 23771897 AAGGCGCAGGAGCCGAGCCGAGCCGAGGAUCCUUUUACAAGUCCC----GGCUUAGACGCCUGGGCCAAAAAAGGCGACGACGUUGUCGCCAACU ..(((.((((.......(..(((((.((((..........)))))----))))..)...)))).)))......(((((((....))))))).... ( -39.60) >DroPse_CAF1 70877 89 - 1 ----AGAAGGAGCCAGGCCAG--AGUGGAUCCUUUUAUAAGAGUCUGCUGGCUUAGACGCCUGGACCAAAAAAGGCGACGACGUUGUCGCCAACU ----....((..((((((...--((..((..(((....)))..))..))(.......))))))).))......(((((((....))))))).... ( -32.50) >DroSec_CAF1 81835 91 - 1 AAGGCGCAGGAGCCGAACCAAGCCGAGGAUCCUUUUACAAGUCCC----GGCUUAGACGCCUGGGCCAAAAAAGGCGACGACGUUGUCGCCAACU ..(((.((((.(.(.....((((((.((((..........)))))----))))).).).)))).)))......(((((((....))))))).... ( -38.00) >DroEre_CAF1 81832 84 - 1 -AGGCGCAGGAGCCG------GCCGAGCAUCCUUUUACAAGGCCC----GGCUUAGACGCCUGGGCCAAAAAAGGCGACGGCGUUGUCGCCAACU -.(((((((..((((------..((.....((((((....(((((----(((......))).)))))..)))))))).)))).))).)))).... ( -37.60) >DroYak_CAF1 85139 90 - 1 -AGGCGCAGGAGCCGAUCCGAGCCGAGGAUCCUUUUACAAGUCCC----GGCUAAGACGCCUGGGCCAAAAAAGGCGACGACGUUGUCGCCAACU -.(((.((((...((.((..(((((.((((..........)))))----))))..)))))))).)))......(((((((....))))))).... ( -40.10) >DroPer_CAF1 71018 89 - 1 ----AGAAGGAGCCAGGCCAG--AGUGGAUCCUUUUAUAAGACUCGGCUGGCUUAGACGCCUGGACCAAAAAAGGCGACGACGUUGUCGCCAACU ----....((..((((((...--(((.((..(((....)))..)).)))(.......))))))).))......(((((((....))))))).... ( -29.50) >consensus _AGGCGCAGGAGCCGAGCCAAGCCGAGGAUCCUUUUACAAGUCCC____GGCUUAGACGCCUGGGCCAAAAAAGGCGACGACGUUGUCGCCAACU .......((((.((............)).))))................(((((((....)))))))......(((((((....))))))).... (-23.52 = -24.18 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:39 2006