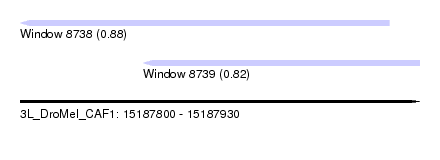
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,187,800 – 15,187,930 |
| Length | 130 |
| Max. P | 0.877689 |

| Location | 15,187,800 – 15,187,920 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -29.41 |
| Energy contribution | -29.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15187800 120 - 23771897 CUCUCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA ...((((((...((((..((((((((((((((((........)))))))))).......((((((........))))))......))))))..))))...))..))))............ ( -31.20) >DroVir_CAF1 66388 118 - 1 --GGCAGGCAACGUUGCGUGUCUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA --.((((....)(((((((((((.((((((((((........)))))))))).......((((......))))))))))..((.....))..........)))))........))).... ( -33.50) >DroGri_CAF1 72454 118 - 1 --GACAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA --(((((((...((((..((((((((((((((((........)))))))))).......((((((........))))))......))))))..))))...))..))).)).......... ( -31.10) >DroSec_CAF1 77383 120 - 1 CUCUCAGGCAACGUUGCGUGUUUGGCCGACUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA ...((((((...((((..((((((((((((((((........)))))))))).......((((((........))))))......))))))..))))...))..))))............ ( -31.70) >DroMoj_CAF1 70807 120 - 1 CAGGCAGGCAACGUUGCGUGUUUGGUCGGCUUUGACCUAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA ((((((.(((....))).))))))..((((((((........))))))))(((((((.(((((((((((((.((((.......)))).)))))....))))).))).)..)))))).... ( -30.40) >DroPer_CAF1 67317 120 - 1 CCGGCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA ...((((....)(((((.((((((((((((((((........)))))))))).......((((((........))))))......)))))).........)))))........))).... ( -32.50) >consensus C_GGCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACAAGUUUCAAGCAAUCAACAAAGCGACUGAUCAAAUGCAAGA ....(((((...((((..((((((((((((((((........)))))))))).......((((((........))))))......))))))..))))...))..)))............. (-29.41 = -29.30 + -0.11)


| Location | 15,187,840 – 15,187,930 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.81 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.25 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15187840 90 - 23771897 ---------------------------AACUCGGAAACUCUCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ---------------------------...((((((.(...((.(((....))).))...)((((((((((........)))))))))).))))))((((((........)))))). ( -24.70) >DroVir_CAF1 66428 78 - 1 ---------------------------------------GGCAGGCAACGUUGCGUGUCUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ---------------------------------------((((.(((....))).))))..((((((((((........)))))))))).......((((((........)))))). ( -25.80) >DroPse_CAF1 67083 117 - 1 CCCAAACCCAAACCCAAUCCCAAUCCCAAUCCGAGAUCCGGCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ..((((..((((((((((............(((.....)))..(....))))).).)))))((((((((((........))))))))))..)))).((((((........)))))). ( -27.90) >DroGri_CAF1 72494 78 - 1 ---------------------------------------GACAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ---------------------------------------((((.(((....))).))))..((((((((((........)))))))))).......((((((........)))))). ( -23.70) >DroAna_CAF1 72378 82 - 1 ---------------------------A--------ACUCUCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ---------------------------.--------.(...((.(((....))).))...)((((((((((........)))))))))).......((((((........)))))). ( -21.80) >DroPer_CAF1 67357 111 - 1 CCCAAA------CCCAAUCCCAAUCCCAAUCCGAGAUCCGGCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ..((((------((((((............(((.....)))..(....))))).).)))))((((((((((........)))))))))).......((((((........)))))). ( -27.00) >consensus ___________________________A_________C_GGCAGGCAACGUUGCGUGUUUGGCCGGCUUUGACCCAAAAUAAAGUUGGCAUUUUGAGUGUUUUAUUGCAUGGACACA ............................................(((....)))((((((.((((((((((........)))))))))).......((((......)))))))))). (-22.39 = -22.25 + -0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:32 2006