| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,181,237 – 15,181,377 |
| Length | 140 |
| Max. P | 0.869102 |

| Location | 15,181,237 – 15,181,338 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.91 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15181237 101 - 23771897 UUUUCAGUUACCAACUGGGGCUAUAUCCCCCCAUUCCCCCCGGCUGUCCAAUCCCCCCAGAUAUCGAUGGUAAUACCGCCGGGUAAUUUUCGUACCUACUG ....(((((((....(((((........))))).....((((((.(((..(((......)))...)))((.....))))))))........)))...)))) ( -24.70) >DroSec_CAF1 70847 86 - 1 UUUUCAGUUACCAACUGGGGCUAUAUCCC---------------UGCCCGAUCCCCCCAGAUAUCGAUGGUAAUACCGCCGGGUAAUUUUCGUACCUGCUG .....(((((((..((((((....(((..---------------.....)))..))))))....((.(((.....))).)))))))))............. ( -20.90) >DroSim_CAF1 72254 86 - 1 UUUUCAGUUACCAACUGGGGCUAUAUCCC---------------UGCCCGAUCCCCCCAGAUAUCGAUGGUAAUACCGCCGGGUAAUUUUCGUACCUGCUG .....(((((((..((((((....(((..---------------.....)))..))))))....((.(((.....))).)))))))))............. ( -20.90) >DroYak_CAF1 73754 97 - 1 UUUUCAGUUACCAACUGGGGCUAUAUCCC--CAUUCCCCCUU--UACGAGGUUCCUCCAGAUAUCGGUGGUAAUACCGCCGGGUAAUUUUCGUACCUGCUG .....(((((((...(((((......)))--)).........--...(((....))).......((((((.....)))))))))))))............. ( -27.10) >consensus UUUUCAGUUACCAACUGGGGCUAUAUCCC_______________UGCCCGAUCCCCCCAGAUAUCGAUGGUAAUACCGCCGGGUAAUUUUCGUACCUGCUG .....(((((((..((((((..((..........................))..))))))....((.(((.....))).)))))))))............. (-18.50 = -18.12 + -0.38)

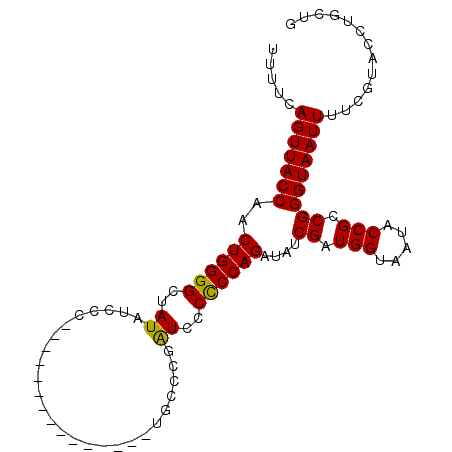

| Location | 15,181,258 – 15,181,377 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15181258 119 + 23771897 GGCGGUAUUACCAUCGAUAUCUGGGGGGAUUGGACAGCCGGGGGGAAUGGGGGGAUAUAGCCCCAGUUGGUAACUGAAAAUAUGAGUAGGCUAAAGGUUUAUCAGCGCCAUAAAUAAGA ((((...((((((.(....(((((..(.......)..))))).....(((((........)))))).))))))((((((((...((....))....)))).)))))))).......... ( -32.50) >DroSec_CAF1 70868 104 + 1 GGCGGUAUUACCAUCGAUAUCUGGGGGGAUCGGGCA---------------GGGAUAUAGCCCCAGUUGGUAACUGAAAAUAUGAGUAGGCUAAAGGUUUAUCAGCGCCAUAAAUAAGA ((((...((((((.......((((((..(((.....---------------..)))....)))))).))))))((((((((...((....))....)))).)))))))).......... ( -28.50) >DroSim_CAF1 72275 104 + 1 GGCGGUAUUACCAUCGAUAUCUGGGGGGAUCGGGCA---------------GGGAUAUAGCCCCAGUUGGUAACUGAAAAUAUGAGUAGGCUAAAGGUUUAUCAGCGCCAUAAAUAAGA ((((...((((((.......((((((..(((.....---------------..)))....)))))).))))))((((((((...((....))....)))).)))))))).......... ( -28.50) >DroYak_CAF1 73775 115 + 1 GGCGGUAUUACCACCGAUAUCUGGAGGAACCUCGUA--AAGGGGGAAUG--GGGAUAUAGCCCCAGUUGGUAACUGAAAAUAUGAGUAGGCUAAAGGUUUAUCAGUGCCAUAAAUAAAA (((((.....))((((((...........((((...--..))))...((--(((......))))))))))).(((((((((...((....))....)))).)))))))).......... ( -32.90) >consensus GGCGGUAUUACCAUCGAUAUCUGGGGGGAUCGGGCA_______________GGGAUAUAGCCCCAGUUGGUAACUGAAAAUAUGAGUAGGCUAAAGGUUUAUCAGCGCCAUAAAUAAGA ((((...((((((.(....(((((.....))))).................(((.......))).).))))))((((((((...((....))....)))).)))))))).......... (-23.30 = -23.05 + -0.25)

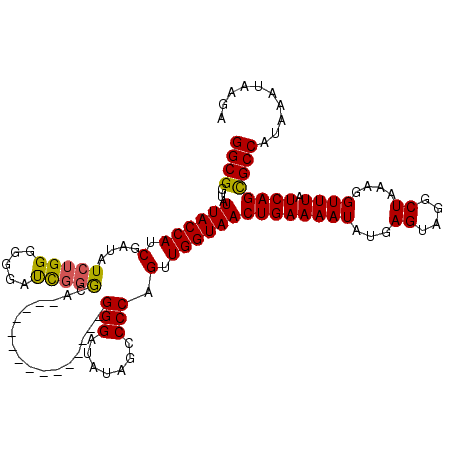

| Location | 15,181,258 – 15,181,377 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.04 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15181258 119 - 23771897 UCUUAUUUAUGGCGCUGAUAAACCUUUAGCCUACUCAUAUUUUCAGUUACCAACUGGGGCUAUAUCCCCCCAUUCCCCCCGGCUGUCCAAUCCCCCCAGAUAUCGAUGGUAAUACCGCC ..........((((((((.......)))))...............(((((((..(((((........))))).......(((.((((...........))))))).)))))))...))) ( -25.30) >DroSec_CAF1 70868 104 - 1 UCUUAUUUAUGGCGCUGAUAAACCUUUAGCCUACUCAUAUUUUCAGUUACCAACUGGGGCUAUAUCCC---------------UGCCCGAUCCCCCCAGAUAUCGAUGGUAAUACCGCC ..........((((((((.......)))))...............(((((((....((((........---------------.)))).(((......))).....)))))))...))) ( -22.90) >DroSim_CAF1 72275 104 - 1 UCUUAUUUAUGGCGCUGAUAAACCUUUAGCCUACUCAUAUUUUCAGUUACCAACUGGGGCUAUAUCCC---------------UGCCCGAUCCCCCCAGAUAUCGAUGGUAAUACCGCC ..........((((((((.......)))))...............(((((((....((((........---------------.)))).(((......))).....)))))))...))) ( -22.90) >DroYak_CAF1 73775 115 - 1 UUUUAUUUAUGGCACUGAUAAACCUUUAGCCUACUCAUAUUUUCAGUUACCAACUGGGGCUAUAUCCC--CAUUCCCCCUU--UACGAGGUUCCUCCAGAUAUCGGUGGUAAUACCGCC ..........(((.((((.......)))).(((((.((((((..((..(((...(((((......)))--)).((......--...)))))..))..)))))).))))).......))) ( -23.40) >consensus UCUUAUUUAUGGCGCUGAUAAACCUUUAGCCUACUCAUAUUUUCAGUUACCAACUGGGGCUAUAUCCC_______________UGCCCGAUCCCCCCAGAUAUCGAUGGUAAUACCGCC ..........((((((((.......)))))...............(((((((.((((((..((..........................))..)))))).......)))))))...))) (-19.17 = -19.04 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:30 2006