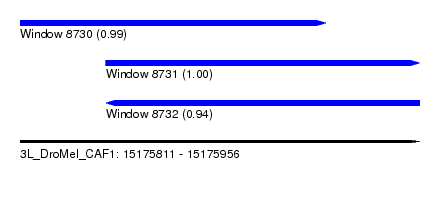
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,175,811 – 15,175,956 |
| Length | 145 |
| Max. P | 0.998515 |

| Location | 15,175,811 – 15,175,922 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -40.30 |
| Energy contribution | -40.10 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15175811 111 + 23771897 UCUUGAGGCCUAAGAACCGGAGACGUGACGCUCUCCUCAGAACCUCAUCGACGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUGGCUGGCGCCUCAGAACGGAGCGA ..((((((((((.(((..(((((((...)).)))))..((((((.((((..(((....)))...)))).)))))))))...((.....))))).))))))).......... ( -37.50) >DroSec_CAF1 65484 111 + 1 UCUUGAGGCCUAAGAACCGGAGACGUGACGCUCUCCUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAACGCUUUUCGCUGGCGCCUCAGAACGGAGCGA ..((((((((((.(((..(((((((...)).)))))..((((((.(((((((((....))))).)))).))))))..........)))..))).))))))).......... ( -42.70) >DroSim_CAF1 66835 111 + 1 UCUUGAGGCCUAAGAACCGGAGACGUGACGCUCUCCUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAACGCUUUUCGCUGGCGCCUCAGAACGGAGCGA ..((((((((((.(((..(((((((...)).)))))..((((((.(((((((((....))))).)))).))))))..........)))..))).))))))).......... ( -42.70) >DroEre_CAF1 66104 111 + 1 UCUUGAGGCCUAAGAACCGGAGACGUGACGCUCUCUGCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUCGCUGGCGCCUCAGAACGGAGCGA ..((((((((((.(((..(....)((((.((.....))((((((.(((((((((....))))).)))).))))))....))))..)))..))).))))))).......... ( -43.00) >DroYak_CAF1 68186 111 + 1 UCUUGAGGCCUAAGAACCGGAGACGUGACGCUCUCUUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUCGCUGGCGCCUCGGAACGGAGCGA (((((.....))))).(((....)).).((((((.(((((((((.(((((((((....))))).)))).)))))).....((((.......)))).....))).)))))). ( -42.40) >consensus UCUUGAGGCCUAAGAACCGGAGACGUGACGCUCUCCUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUCGCUGGCGCCUCAGAACGGAGCGA (((.((((((((.(((..(((((((...)).)))))..((((((.(((((((((....))))).)))).)))))))))...((.....))))).))))))))......... (-40.30 = -40.10 + -0.20)

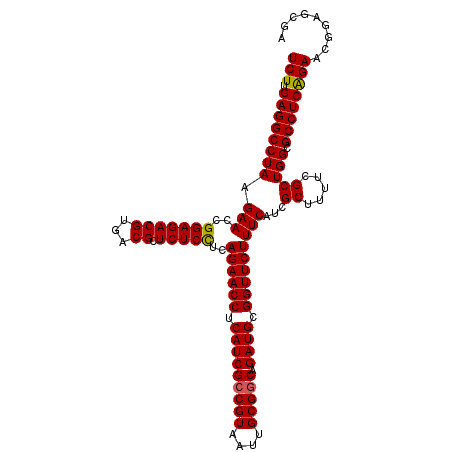
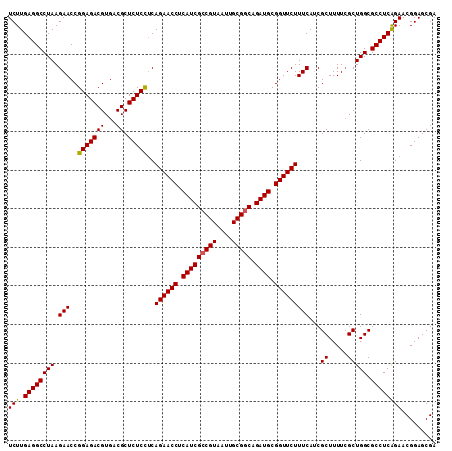
| Location | 15,175,842 – 15,175,956 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -36.18 |
| Energy contribution | -36.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15175842 114 + 23771897 UCUCCUCAGAACCUCAUCGACGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUGGCUGGCGCCUCAGAACGGAGCGACGUGCUCUUUCUCACCUGGCUUCACCUGGCCCCA .......((((((.((((..(((....)))...)))).))))))......(((...((.(((.(((..((((.(((((.....))))))))).....))).))))).))).... ( -35.60) >DroSec_CAF1 65515 114 + 1 UCUCCUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAACGCUUUUCGCUGGCGCCUCAGAACGGAGCGACGUGCUCUUUCUCACCUGGCUUCACCUGGCCCCA .......((((((.(((((((((....))))).)))).)))))).....((((.......))))....((((.(((((.....))))))))).....((((......))))... ( -37.80) >DroSim_CAF1 66866 114 + 1 UCUCCUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAACGCUUUUCGCUGGCGCCUCAGAACGGAGCGACGUGCUCUUUCUCACCUGGCUUCACCUGGCCCCA .......((((((.(((((((((....))))).)))).)))))).....((((.......))))....((((.(((((.....))))))))).....((((......))))... ( -37.80) >DroEre_CAF1 66135 112 + 1 UCUCUGCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUCGCUGGCGCCUCAGAACGGAGCGACGUGCUCUUUCUCACCUGGCUUCACCUGGCCC-- .....((((((((.(((((((((....))))).)))).))))).....(((((((...((((....))))...)))))))..)))............((((......)))).-- ( -39.60) >DroYak_CAF1 68217 114 + 1 UCUCUUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUCGCUGGCGCCUCGGAACGGAGCGACGUGCUCUUUCUCACCUGGCUUCACCUGGCCCCA .......((((((.(((((((((....))))).)))).))))))......((.....)).((.(((..((...(((((.(.(((........))).).))))).)).))).)). ( -39.70) >consensus UCUCCUCAGAACCUCAUCGCCGUAAUUGCGGCAGAUGCGGUUCUUUCAUCGCUUUUCGCUGGCGCCUCAGAACGGAGCGACGUGCUCUUUCUCACCUGGCUUCACCUGGCCCCA .......((((((.(((((((((....))))).)))).)))))).....((((.......))))....((((.(((((.....))))))))).....((((......))))... (-36.18 = -36.22 + 0.04)

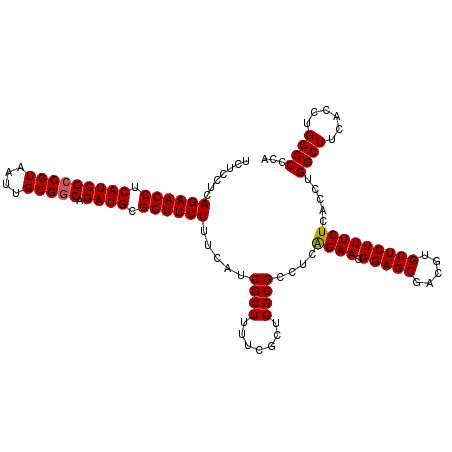
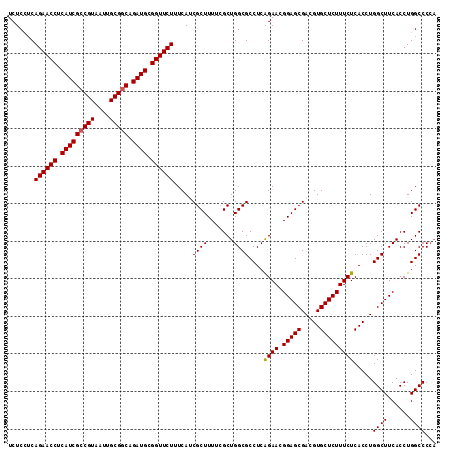
| Location | 15,175,842 – 15,175,956 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -38.84 |
| Energy contribution | -39.64 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
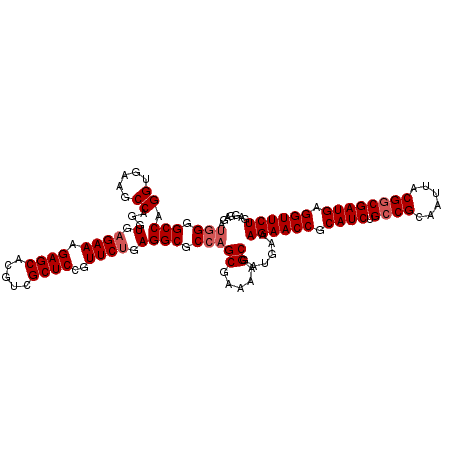
>3L_DroMel_CAF1 15175842 114 - 23771897 UGGGGCCAGGUGAAGCCAGGUGAGAAAGAGCACGUCGCUCCGUUCUGAGGCGCCAGCCAAAAGCGAUGAAAGAACCGCAUCUGCCGCAAUUACGUCGAUGAGGUUCUGAGGAGA (((.(((.((.....))...(.((((.((((.....))))..)))).)))).)))((.....))......((((((.((((.(.((......)).))))).))))))....... ( -35.00) >DroSec_CAF1 65515 114 - 1 UGGGGCCAGGUGAAGCCAGGUGAGAAAGAGCACGUCGCUCCGUUCUGAGGCGCCAGCGAAAAGCGUUGAAAGAACCGCAUCUGCCGCAAUUACGGCGAUGAGGUUCUGAGGAGA (((.(((.((.....))...(.((((.((((.....))))..)))).)))).)))((.....))......((((((.((((.((((......)))))))).))))))....... ( -43.30) >DroSim_CAF1 66866 114 - 1 UGGGGCCAGGUGAAGCCAGGUGAGAAAGAGCACGUCGCUCCGUUCUGAGGCGCCAGCGAAAAGCGUUGAAAGAACCGCAUCUGCCGCAAUUACGGCGAUGAGGUUCUGAGGAGA (((.(((.((.....))...(.((((.((((.....))))..)))).)))).)))((.....))......((((((.((((.((((......)))))))).))))))....... ( -43.30) >DroEre_CAF1 66135 112 - 1 --GGGCCAGGUGAAGCCAGGUGAGAAAGAGCACGUCGCUCCGUUCUGAGGCGCCAGCGAAAAGCGAUGAAAGAACCGCAUCUGCCGCAAUUACGGCGAUGAGGUUCUGCAGAGA --(((((.((.....))...(.((((.((((.....))))..)))).)))).)).((.....))......((((((.((((.((((......)))))))).))))))....... ( -40.40) >DroYak_CAF1 68217 114 - 1 UGGGGCCAGGUGAAGCCAGGUGAGAAAGAGCACGUCGCUCCGUUCCGAGGCGCCAGCGAAAAGCGAUGAAAGAACCGCAUCUGCCGCAAUUACGGCGAUGAGGUUCUGAAGAGA (((.(((.((.....))..(.(((...((((.....))))..))))..))).)))((.....))......((((((.((((.((((......)))))))).))))))....... ( -41.70) >consensus UGGGGCCAGGUGAAGCCAGGUGAGAAAGAGCACGUCGCUCCGUUCUGAGGCGCCAGCGAAAAGCGAUGAAAGAACCGCAUCUGCCGCAAUUACGGCGAUGAGGUUCUGAGGAGA (((.(((.((.....))...(.((((.((((.....))))..)))).)))).)))((.....))......((((((.((((.((((......)))))))).))))))....... (-38.84 = -39.64 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:25 2006