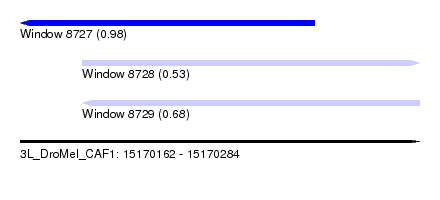
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,170,162 – 15,170,284 |
| Length | 122 |
| Max. P | 0.984902 |

| Location | 15,170,162 – 15,170,252 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.29 |
| Mean single sequence MFE | -18.71 |
| Consensus MFE | -13.16 |
| Energy contribution | -14.47 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15170162 90 - 23771897 AAAUGGCUUUAGCUUAAGCUCUACAAAAACAGUUUAUAGUAUAACCAUUUCCGGAAAGUAUCCAAUUUGAA-----------------------AACU-A----AAAACUUCUUAAUU ((((((.((..(((((((((..........)))))).)))..))))))))..(((.....)))........-----------------------....-.----.............. ( -9.40) >DroSec_CAF1 59875 113 - 1 AAAUGGUUUUAGCUUAAGCUCUACAAAAAGAGUUUAUAGUAAAACCAUUUCCGGUAAGAACCCAAUUUGAAAUACCUCGUUAAUCUGUACAGAAAACU-A----AAAACUUUUUUAAA (((((((((((.((((((((((......)))))))).)))))))))))))..((((................))))..(((..(((....))).))).-.----.............. ( -21.09) >DroSim_CAF1 61139 114 - 1 AAAUGGUUUUAGCUUAAGCUCUACAAAAAGAGUUUAUAGCAUAACCAUUUCCGGAAAGAUUCCAAUUUGAAAUACCUAUUUAAUCUGCACAGAAAUCUGA----AAAACUUCAUAAAA ((((((((...(((((((((((......)))))))).)))..))))))))..(((.....)))..........................(((....))).----.............. ( -23.00) >DroEre_CAF1 60884 115 - 1 AAAUGGUUUAAGCUUAAGCUCUACCAGAAAC-CUUAAAGCAUACCCAUUUCCGGAAAGUAUCCACUUUUAAUUACCUUCUAUAUCUUUGCGGAAAACU-ACCCGAAAG-UGCUUAGAA ...((((...(((....)))..)))).....-....(((((......((((((.((((..........................)))).))))))...-...(....)-))))).... ( -21.37) >consensus AAAUGGUUUUAGCUUAAGCUCUACAAAAAGAGUUUAUAGCAUAACCAUUUCCGGAAAGAAUCCAAUUUGAAAUACCU__UUAAUCUGUACAGAAAACU_A____AAAACUUCUUAAAA ((((((((...(((((((((((......)))))))).)))..))))))))..(((.....)))....................................................... (-13.16 = -14.47 + 1.31)

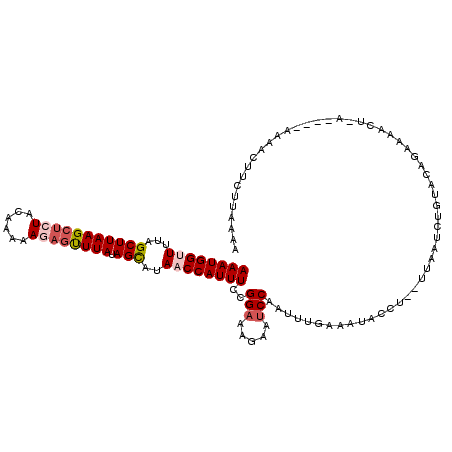

| Location | 15,170,181 – 15,170,284 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -14.68 |
| Energy contribution | -15.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15170181 103 + 23771897 -------UUCAAAUUGGAUACUUUCCGGAAAUGGUUAUACUAUAAACUGUUUUUGUAGAGCUUAAGCUAAAGCCAUUUAAACGCAAAUAUAAUCACUUUGUUUUUUUGCC -------......(((((.....)))))((((((((...(((((((.....)))))))(((....)))..))))))))....(((((.((((.....))))...))))). ( -22.10) >DroSec_CAF1 59910 104 + 1 GAGGUAUUUCAAAUUGGGUUCUUACCGGAAAUGGUUUUACUAUAAACUCUUUUUGUAGAGCUUAAGCUAAAACCAUUUAAAUGCAA-----AUCACACUGUUU-UUUGCC ((.((((((.......(((....)))..(((((((((((((.(((.((((......)))).))))).)))))))))))))))))..-----.)).........-...... ( -22.70) >DroSim_CAF1 61175 109 + 1 UAGGUAUUUCAAAUUGGAAUCUUUCCGGAAAUGGUUAUGCUAUAAACUCUUUUUGUAGAGCUUAAGCUAAAACCAUUUAAAUGCAAAUAUAAUCACACUGUUU-UUUGCC ...((((((....(((((.....)))))((((((((..(((.(((.((((......)))).))))))...))))))))))))))...................-...... ( -21.60) >DroEre_CAF1 60922 108 + 1 AAGGUAAUUAAAAGUGGAUACUUUCCGGAAAUGGGUAUGCUUUAAG-GUUUCUGGUAGAGCUUAAGCUUAAACCAUUUAAAUGCAAAUAUAAUCACAUUGUUU-UUUGCC ..(((((..((.(((((((....((((....))))..(((((((((-.....((((.((((....))))..)))))))))).)))......))).)))).)).-.))))) ( -21.70) >consensus _AGGUAUUUCAAAUUGGAUACUUUCCGGAAAUGGUUAUACUAUAAACUCUUUUUGUAGAGCUUAAGCUAAAACCAUUUAAAUGCAAAUAUAAUCACACUGUUU_UUUGCC ..............((((.....)))).((((((((...(((((((.....)))))))(((....)))..))))))))....(((((...(((......)))..))))). (-14.68 = -15.68 + 1.00)

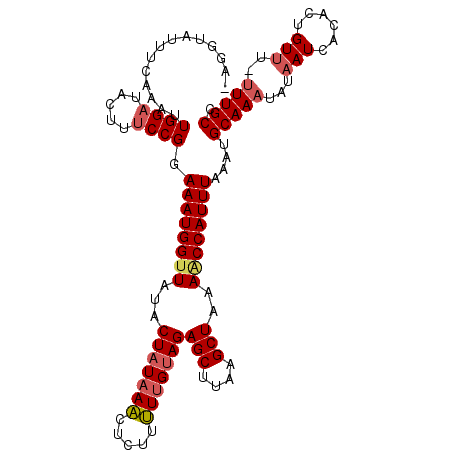
| Location | 15,170,181 – 15,170,284 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -16.79 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15170181 103 - 23771897 GGCAAAAAAACAAAGUGAUUAUAUUUGCGUUUAAAUGGCUUUAGCUUAAGCUCUACAAAAACAGUUUAUAGUAUAACCAUUUCCGGAAAGUAUCCAAUUUGAA------- ..((((......(((((.(((((((.((((((....(((((......)))))......)))).))....))))))).)))))..(((.....)))..))))..------- ( -14.80) >DroSec_CAF1 59910 104 - 1 GGCAAA-AAACAGUGUGAU-----UUGCAUUUAAAUGGUUUUAGCUUAAGCUCUACAAAAAGAGUUUAUAGUAAAACCAUUUCCGGUAAGAACCCAAUUUGAAAUACCUC .(((((-..((...))..)-----))))....(((((((((((.((((((((((......)))))))).)))))))))))))..((((................)))).. ( -24.29) >DroSim_CAF1 61175 109 - 1 GGCAAA-AAACAGUGUGAUUAUAUUUGCAUUUAAAUGGUUUUAGCUUAAGCUCUACAAAAAGAGUUUAUAGCAUAACCAUUUCCGGAAAGAUUCCAAUUUGAAAUACCUA ((....-....((((..(......)..)))).((((((((...(((((((((((......)))))))).)))..))))))))))(((.....)))............... ( -25.40) >DroEre_CAF1 60922 108 - 1 GGCAAA-AAACAAUGUGAUUAUAUUUGCAUUUAAAUGGUUUAAGCUUAAGCUCUACCAGAAAC-CUUAAAGCAUACCCAUUUCCGGAAAGUAUCCACUUUUAAUUACCUU ......-.......(((((((....(((.(((((.((((...(((....)))..)))).....-.))))))))...........(((.....))).....)))))))... ( -16.00) >consensus GGCAAA_AAACAAUGUGAUUAUAUUUGCAUUUAAAUGGUUUUAGCUUAAGCUCUACAAAAAGAGUUUAUAGCAUAACCAUUUCCGGAAAGAAUCCAAUUUGAAAUACCU_ ((.........((((..(......)..)))).((((((((...(((((((((((......)))))))).)))..))))))))))(((.....)))............... (-16.79 = -18.10 + 1.31)

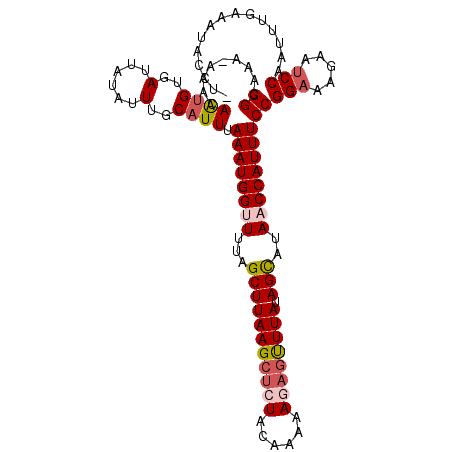
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:22 2006