
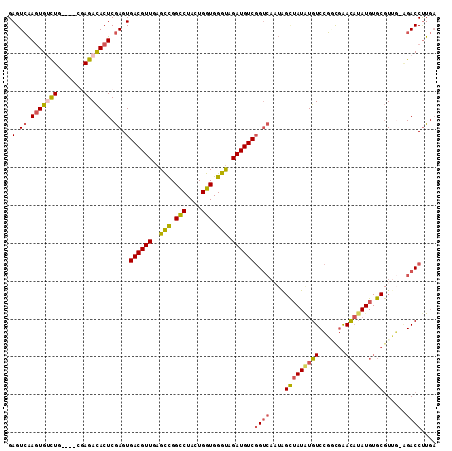
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,163,548 – 15,163,661 |
| Length | 113 |
| Max. P | 0.861987 |

| Location | 15,163,548 – 15,163,661 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -26.73 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
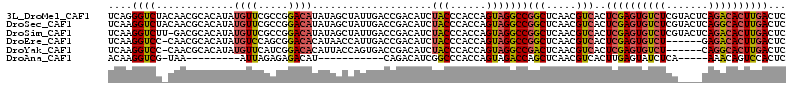
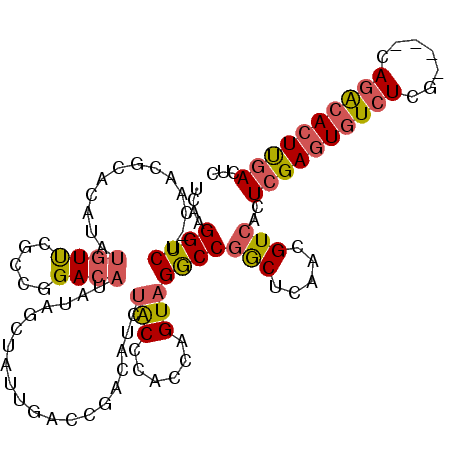
>3L_DroMel_CAF1 15163548 113 + 23771897 GAGUCAAGUGUCUGAGUACGAGACACUCGAGUGACGUUGAGCCGGCCUACUGGUGGGUAGAUGUCGGUCAAUAGCUAUAUGUCCGGCGAACAUAUGUGCGUUGUAGACCCUGA .(.((.(((((((.......))))))).)).)((((((..(((.(((....))).))).))))))((((.((((((((((((.......)))))))...))))).)))).... ( -39.90) >DroSec_CAF1 53249 113 + 1 GAGUCAAGUGCCUGAGUACGAGACACUCGAGUGACGUUGAGCCGGCCUACUGGUGGGUAGAUGUCGGUCAAUAGCUAUAUGUCCGGCGAACAUAUGUGCGUUGUAGACCUUGA ..(((..((((....))))..)))..((((..((((((..(((.(((....))).))).))))))((((.((((((((((((.......)))))))...))))).)))))))) ( -37.90) >DroSim_CAF1 54530 112 + 1 GAGUCAAGUGUCUGAGUACGAGACACUCGAGUGACGUUGAGCCGGCCUACUGGUGGGUAGAUGUCGGUCAAUAGCUAUAUGUCCGGCGAACAUAUGUGCGUC-AAGACCUUGA ...(((((.((((((((.......))))((.(((((((..(((.(((....))).))).))))))).))....(((((((((.......))))))).))...-.))))))))) ( -39.80) >DroEre_CAF1 54287 106 + 1 GAGUCAAGUGUCUC------AGACACUCGAGUGACGUUGAGCCGGCCUACUGGUGGGUAGAUGUCGGUCAAUGGUUAUGUGUCCGCUGGACAUAUGUGCGUUG-GGACCUUGA ...(((((.(((((------((.(((..((.(((((((..(((.(((....))).))).))))))).)).......((((((((...)))))))))))..)))-))))))))) ( -45.70) >DroYak_CAF1 54595 106 + 1 GAGUCAAGUGCCUG------AGACACUCGAGUGACGUUGAGUCGGCCUACUGGUGGGUAGAUGUCGGUCACUGGUAAUGUGUCCGAUGAACAUAUGUGCGUUG-GGACCUUGA ...(((((..((..------.(((((.(.((((((.....(((.(((((....))))).)))....)))))).)....)))))(((((.((....)).)))))-))..))))) ( -36.60) >DroAna_CAF1 49507 87 + 1 GAGUGGACUGUUU-----UGAGAUACUCAAGUGACGUUGAGCUGGUCUACUGGUGGGCCGAUGUCUG-----------AUGUCUCUCUAAU---------UUA-CGACCUUGU (((.((((...((-----((((...)))))).(((((((.(((..((....))..))))))))))..-----------..)))))))....---------...-......... ( -22.90) >consensus GAGUCAAGUGUCUG____CGAGACACUCGAGUGACGUUGAGCCGGCCUACUGGUGGGUAGAUGUCGGUCAAUAGCUAUAUGUCCGGCGAACAUAUGUGCGUUG_AGACCUUGA .(.((.(((((((.......))))))).)).)((((((..(((.(((....))).))).))))))((((....(((((((((.......))))))).))......)))).... (-26.73 = -27.30 + 0.57)


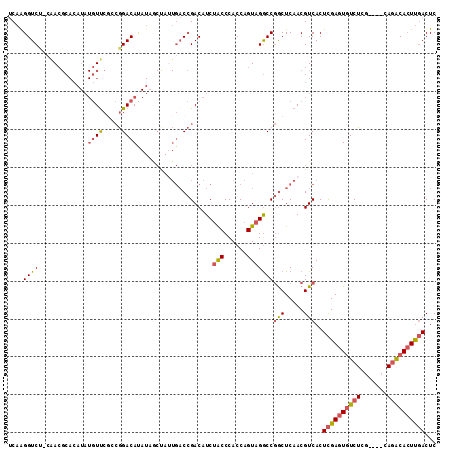
| Location | 15,163,548 – 15,163,661 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15163548 113 - 23771897 UCAGGGUCUACAACGCACAUAUGUUCGCCGGACAUAUAGCUAUUGACCGACAUCUACCCACCAGUAGGCCGGCUCAACGUCACUCGAGUGUCUCGUACUCAGACACUUGACUC ....(((((((...((..(((((((.....))))))).))....((......)).........)))))))(((.....)))..((((((((((.......))))))))))... ( -31.60) >DroSec_CAF1 53249 113 - 1 UCAAGGUCUACAACGCACAUAUGUUCGCCGGACAUAUAGCUAUUGACCGACAUCUACCCACCAGUAGGCCGGCUCAACGUCACUCGAGUGUCUCGUACUCAGGCACUUGACUC ....(((((((...((..(((((((.....))))))).))....((......)).........)))))))(((.....)))..((((((((((.......))))))))))... ( -31.20) >DroSim_CAF1 54530 112 - 1 UCAAGGUCUU-GACGCACAUAUGUUCGCCGGACAUAUAGCUAUUGACCGACAUCUACCCACCAGUAGGCCGGCUCAACGUCACUCGAGUGUCUCGUACUCAGACACUUGACUC ((((((((((-(((((..(((((((.....))))))).))..(((((((...(((((......))))).))).)))).))))...(((((.....))))))))).)))))... ( -34.00) >DroEre_CAF1 54287 106 - 1 UCAAGGUCC-CAACGCACAUAUGUCCAGCGGACACAUAACCAUUGACCGACAUCUACCCACCAGUAGGCCGGCUCAACGUCACUCGAGUGUCU------GAGACACUUGACUC ((((((((.-((.(((...........)))(((((.......(((((((...(((((......))))).))).))))((.....)).))))))------).))).)))))... ( -28.10) >DroYak_CAF1 54595 106 - 1 UCAAGGUCC-CAACGCACAUAUGUUCAUCGGACACAUUACCAGUGACCGACAUCUACCCACCAGUAGGCCGACUCAACGUCACUCGAGUGUCU------CAGGCACUUGACUC ....(((..-.................((((.(((.......))).))))...((((......)))))))(((.....)))..((((((((..------...))))))))... ( -25.80) >DroAna_CAF1 49507 87 - 1 ACAAGGUCG-UAA---------AUUAGAGAGACAU-----------CAGACAUCGGCCCACCAGUAGACCAGCUCAACGUCACUUGAGUAUCUCA-----AAACAGUCCACUC ....((((.-...---------....((......)-----------).......((....))....)))).((((((......))))))......-----............. ( -13.20) >consensus UCAAGGUCU_CAACGCACAUAUGUUCGCCGGACAUAUAGCUAUUGACCGACAUCUACCCACCAGUAGGCCGGCUCAACGUCACUCGAGUGUCUCG____CAGACACUUGACUC ....((((.............((((.....))))....................(((......)))))))(((.....)))..((((((((((.......))))))))))... (-17.01 = -17.43 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:18 2006