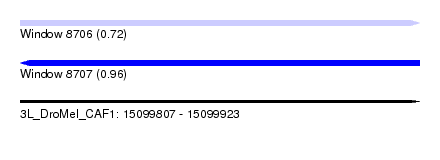
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,099,807 – 15,099,923 |
| Length | 116 |
| Max. P | 0.959062 |

| Location | 15,099,807 – 15,099,923 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15099807 116 + 23771897 GCACUCAGUUGAG-GUUGUAGUCAUUUUG-GGACAGCGAAGUGAAGCCAGCGACUGCUCGUCACGCGACUAACUGCGCCGGCGGUGGCUUUUGCCCCACUCGCUUGUCCGAAUCAGAC ...(((....)))-......(((...((.-(((((((((.(((..((.(((.(((((.((...((((......)))).))))))).)))...))..))))))).))))).))...))) ( -42.40) >DroVir_CAF1 3258 117 + 1 GUACUCAGAUAAAUGUUGAAGUCAUUUUAUGGGCAGCAGAGGCACGCCCGCCUUCACCUGGCACGCGACUAACUGCGUGGG-GAAGGGCUACGUGCACACCGCAUGCCCAACUCAGAC ...((.((..(((((.......)))))..((((((((.(.((((((...(((((..((...((((((......))))))))-..)))))..))))).).).)).)))))).)).)).. ( -40.90) >DroPse_CAF1 8605 116 + 1 UUACUCAGCUGAG-AUUGUAGUCACUUUG-GGAUAGCCGAGUGGAGCCAGCCUCCGCCUGUCACGCGACUAACUGCGUCGGCGGAGCCUUUUGCUCCAUCUGCUUGUCCGAAUCAGAC ...(((....)))-......(((...((.-(((((((.((.((((((.((.(((((((....(((((......))))).))))))).))...)))))))).)).))))).))...))) ( -45.10) >DroYak_CAF1 2869 116 + 1 GCACUCAGUUGAG-GUUGAAGUCAUUUUG-GGACAGCGAAGAGGAGCCAGCUACUGCCCGUCACGCGACUAACUGCGCCGGCAGUGGCUUUUGCCCAACCCGCUUGUCCGAAUCAGAC ...(((....)))-......(((...((.-((((((((....((.((.((((((((((.(...((((......))))).))))))))))...))))....))).))))).))...))) ( -42.20) >DroAna_CAF1 2594 117 + 1 GCACUCAGCAAGG-UUUGUAGUCAUUUUGAGGACAGCAAAGUGGAGCCAGCCAACGCCUGUCACGCGACUAACUGCGCCGGCGACGGCCUCCGCCCCACCUGCUCAUCCGAAUCAGAC ......((((.((-(((((.(((........))).)))).((((((...(((..((((.(...((((......))))).))))..)))))))))...))))))).............. ( -35.30) >DroPer_CAF1 7647 116 + 1 UUACUCAGCUGAG-AUUGUAGUCACUUUG-GGACAGCCGAGUGGAGCCAGCCUCCGCCUGUCACGCGACUAACUGCGUCGGCGGAGCCUUUUGCUCCAUCUGCUUGUCCGAAUCAGAC ...(((....)))-......(((...((.-(((((((.((.((((((.((.(((((((....(((((......))))).))))))).))...)))))))).)).))))).))...))) ( -47.00) >consensus GCACUCAGCUGAG_GUUGUAGUCAUUUUG_GGACAGCAAAGUGGAGCCAGCCACCGCCUGUCACGCGACUAACUGCGCCGGCGGAGGCUUUUGCCCCACCCGCUUGUCCGAAUCAGAC ..............................(((((((...((((.((.((((((((((.....((((......))))..))))))))))...)).))))..)).)))))......... (-28.14 = -28.90 + 0.76)



| Location | 15,099,807 – 15,099,923 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -46.52 |
| Consensus MFE | -40.91 |
| Energy contribution | -40.00 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15099807 116 - 23771897 GUCUGAUUCGGACAAGCGAGUGGGGCAAAAGCCACCGCCGGCGCAGUUAGUCGCGUGACGAGCAGUCGCUGGCUUCACUUCGCUGUCC-CAAAAUGACUACAAC-CUCAACUGAGUGC (((......(((((.(((((((((((....)))...((((((((.(((.(((....))).))).).))))))).)))).)))))))))-......)))......-(((....)))... ( -44.30) >DroVir_CAF1 3258 117 - 1 GUCUGAGUUGGGCAUGCGGUGUGCACGUAGCCCUUC-CCCACGCAGUUAGUCGCGUGCCAGGUGAAGGCGGGCGUGCCUCUGCUGCCCAUAAAAUGACUUCAACAUUUAUCUGAGUAC (((.....((((((.((((.(.((((((.(((.(((-(((((((........)))))...)).))))))..))))))).))))))))))......))).................... ( -47.90) >DroPse_CAF1 8605 116 - 1 GUCUGAUUCGGACAAGCAGAUGGAGCAAAAGGCUCCGCCGACGCAGUUAGUCGCGUGACAGGCGGAGGCUGGCUCCACUCGGCUAUCC-CAAAGUGACUACAAU-CUCAGCUGAGUAA (((((...)))))........(((((...((.(((((((.((((........))))....))))))).)).)))))((((((((....-...............-...)))))))).. ( -47.05) >DroYak_CAF1 2869 116 - 1 GUCUGAUUCGGACAAGCGGGUUGGGCAAAAGCCACUGCCGGCGCAGUUAGUCGCGUGACGGGCAGUAGCUGGCUCCUCUUCGCUGUCC-CAAAAUGACUUCAAC-CUCAACUGAGUGC (((......(((((.(((((..((((...(((.((((((.((((.((.....))))).).)))))).))).)).))..))))))))))-......)))......-(((....)))... ( -43.60) >DroAna_CAF1 2594 117 - 1 GUCUGAUUCGGAUGAGCAGGUGGGGCGGAGGCCGUCGCCGGCGCAGUUAGUCGCGUGACAGGCGUUGGCUGGCUCCACUUUGCUGUCCUCAAAAUGACUACAAA-CCUUGCUGAGUGC (((......((((.((((((((((((...(((((.((((.((((........))))....)))).))))).))))))).))))))))).......)))......-............. ( -45.82) >DroPer_CAF1 7647 116 - 1 GUCUGAUUCGGACAAGCAGAUGGAGCAAAAGGCUCCGCCGACGCAGUUAGUCGCGUGACAGGCGGAGGCUGGCUCCACUCGGCUGUCC-CAAAGUGACUACAAU-CUCAGCUGAGUAA (((((...)))))........(((((...((.(((((((.((((........))))....))))))).)).)))))(((((((((...-...............-..))))))))).. ( -50.45) >consensus GUCUGAUUCGGACAAGCAGGUGGGGCAAAAGCCUCCGCCGACGCAGUUAGUCGCGUGACAGGCGGAGGCUGGCUCCACUCCGCUGUCC_CAAAAUGACUACAAC_CUCAGCUGAGUAC (((......(((((.(((((((((((...((((((((((.((((........))))....)))))))))).)))))).)))))))))).......))).................... (-40.91 = -40.00 + -0.91)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:52:01 2006