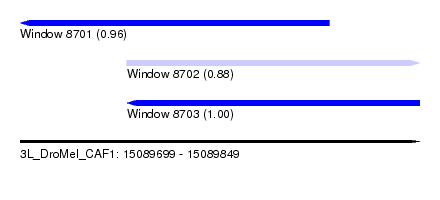
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,089,699 – 15,089,849 |
| Length | 150 |
| Max. P | 0.996769 |

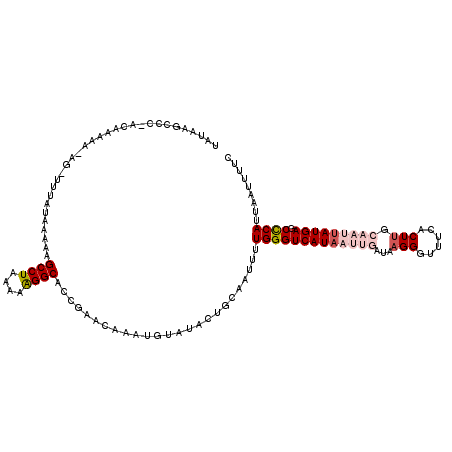
| Location | 15,089,699 – 15,089,815 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -14.63 |
| Energy contribution | -16.48 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
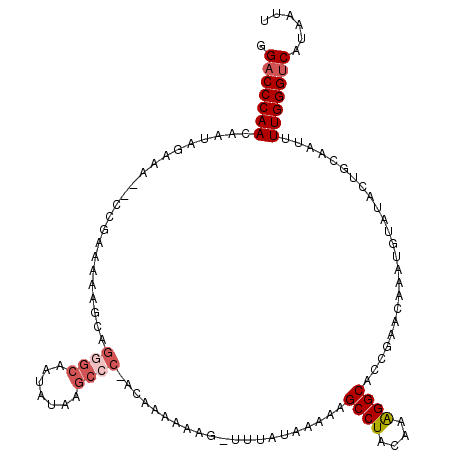
>3L_DroMel_CAF1 15089699 116 - 23771897 UAUAAGCCC-ACAAAAA-AG-UGUAUAAAAAGCCUA-AAAGGCACCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUC .....((.(-((.....-.)-))........(((..-...)))..................)).....(((((((((((((.((((.......))))))))))))).))))......... ( -26.00) >DroSec_CAF1 6982 118 - 1 UAAAAGCCCCACAAAAA-AG-UUUAUAAAAAGCCUACAAAGGCACCGAACAAAUGUAAACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUC .................-((-((((((....((((....))))..........)))))))).......(((((((((((((.((((.......))))))))))))).))))......... ( -25.94) >DroSim_CAF1 7581 118 - 1 UAUAAGCCCCCACAAAA-AG-UUUAUAAAAAGCCUACAAAGGCUCCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUC (((((((..........-.)-))))))...(((((....))))).........(((.....)))....(((((((((((((.((((.......))))))))))))).))))......... ( -26.70) >DroEre_CAF1 6997 101 - 1 UAUAAAACC-ACAAUAA-ACCU-UAAUAAAAGCCUAAAAGGGCACUGAAUAAAUUUACAAUACAAUUUUGGGUCAUAAUUGAUA----------------ACAUGAGCUCAUUAGUUUUC ...(((((.-.......-....-........((((....)))).........................((((((((........----------------..)))).))))...))))). ( -10.50) >DroYak_CAF1 7025 104 - 1 UAUAAAACC-ACAAAAAAACCUUUAUCAAAAGCCUAAAAAGGCAACCAACAAAUGUAUAAUACAACUUUGGGUCAUAAUUGAUAA---------------AUAUGAGCUCAUUAAUUUCC .........-...........((((((((..((((....))))..((((....((((...))))...)))).......)))))))---------------)................... ( -14.80) >consensus UAUAAGCCC_ACAAAAA_AG_UUUAUAAAAAGCCUAAAAAGGCACCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUC ...............................((((....)))).........................(((((((((((((...(((......))).))))))))).))))......... (-14.63 = -16.48 + 1.85)


| Location | 15,089,739 – 15,089,849 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.79 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -18.17 |
| Energy contribution | -19.68 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15089739 110 + 23771897 AAUUAUGACCCAAAAUUGCAGUAUACAUUUGUUCGGUGCCUUU-UAGGCUUUUUAUACA-CUUUUUUGU-GGGCUUAUAUUGCCCUGCUUUUUCGG--UUUCUAUUGUUGGGUCC ......(((((((....((((.......))))..((.(((...-.((((.......(((-......)))-((((.......)))).))))....))--).)).....))))))). ( -25.40) >DroSec_CAF1 7022 112 + 1 AAUUAUGACCCAAAAUUGCAGUUUACAUUUGUUCGGUGCCUUUGUAGGCUUUUUAUAAA-CUUUUUUGUGGGGCUUUUAUUGCACUGCUUUUUCGG--UUUCUAUUGUUGGGUCC ......((((((.....(((((..((.......((((((...((.((((((..((((((-....)))))))))))).))..))))))........)--)....))))))))))). ( -25.46) >DroSim_CAF1 7621 112 + 1 AAUUAUGACCCAAAAUUGCAGUAUACAUUUGUUCGGAGCCUUUGUAGGCUUUUUAUAAA-CUUUUUGUGGGGGCUUAUAUUGCCCUGCUUUUUCGG--UUUUUAUUGUUGGGUCC ......((((((.....((((((.((.(((((..(((((((....)))))))..)))))-.........(((((.......))))).........)--)...)))))))))))). ( -32.90) >DroEre_CAF1 7021 105 + 1 AAUUAUGACCCAAAAUUGUAUUGUAAAUUUAUUCAGUGCCCUUUUAGGCUUUUAUUA-AGGUUUAUUGU-GGUUUUAUAUUGCUUUGCU-------CCCUUCUAUUGUUGGG-CC ...((.(((((((....((((((..........)))))).....((((((((....)-)))))))))).-)))).))....(((..((.-------..........))..))-). ( -15.90) >consensus AAUUAUGACCCAAAAUUGCAGUAUACAUUUGUUCGGUGCCUUUGUAGGCUUUUUAUAAA_CUUUUUUGU_GGGCUUAUAUUGCCCUGCUUUUUCGG__UUUCUAUUGUUGGGUCC ......(((((((........................((((....))))..................(((((((.......)))))))...................))))))). (-18.17 = -19.68 + 1.50)

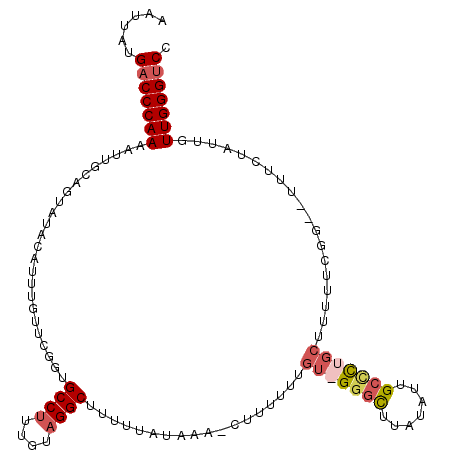
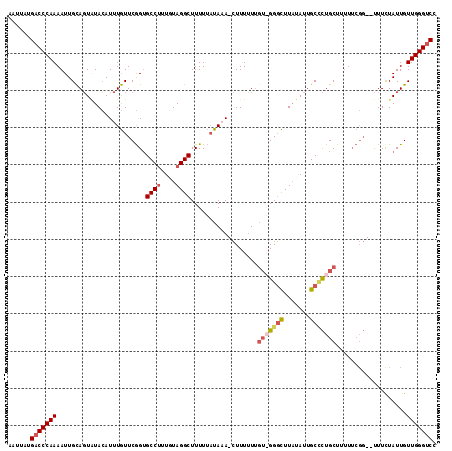
| Location | 15,089,739 – 15,089,849 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.79 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -16.01 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15089739 110 - 23771897 GGACCCAACAAUAGAAA--CCGAAAAAGCAGGGCAAUAUAAGCCC-ACAAAAAAG-UGUAUAAAAAGCCUA-AAAGGCACCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUU .(((((((.....(...--.)......((.((((.......))))-.......((-((((((....(((..-...)))..........))))))))))....)))))))...... ( -26.34) >DroSec_CAF1 7022 112 - 1 GGACCCAACAAUAGAAA--CCGAAAAAGCAGUGCAAUAAAAGCCCCACAAAAAAG-UUUAUAAAAAGCCUACAAAGGCACCGAACAAAUGUAAACUGCAAUUUUGGGUCAUAAUU .(((((((.....(...--.)......((((((((....((((...........)-))).......((((....))))..........)))..)))))....)))))))...... ( -22.30) >DroSim_CAF1 7621 112 - 1 GGACCCAACAAUAAAAA--CCGAAAAAGCAGGGCAAUAUAAGCCCCCACAAAAAG-UUUAUAAAAAGCCUACAAAGGCUCCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUU .((((((((........--..).....((((.(((.(((((((...........)-))))))...(((((....))))).........)))...))))....)))))))...... ( -25.40) >DroEre_CAF1 7021 105 - 1 GG-CCCAACAAUAGAAGGG-------AGCAAAGCAAUAUAAAACC-ACAAUAAACCU-UAAUAAAAGCCUAAAAGGGCACUGAAUAAAUUUACAAUACAAUUUUGGGUCAUAAUU ((-(((((......((((.-------...................-........)))-).......((((....))))........................)))))))...... ( -18.19) >consensus GGACCCAACAAUAGAAA__CCGAAAAAGCAGGGCAAUAUAAGCCC_ACAAAAAAG_UUUAUAAAAAGCCUACAAAGGCACCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUU .(((((((......................((((.......)))).....................((((....))))........................)))))))...... (-16.01 = -17.32 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:57 2006