
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,079,461 – 15,079,551 |
| Length | 90 |
| Max. P | 0.981277 |

| Location | 15,079,461 – 15,079,551 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.77 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15079461 90 + 23771897 CAG------AAGCCAAGGCAUCUGGAAUUUCAAAAGAAGAGGAAAUAAAUGAUCUACUCAAAUGCUUGGGCAGCAAACUGCCAAAGGG----UAAUUAGA (((------(.((....)).))))..............(((...............)))...(((((.(((((....)))))...)))----))...... ( -21.26) >DroSec_CAF1 44481 94 + 1 CAG------AAGCCAAGGCAUCUGGAAUUUCCAAAGAAGAGGAAAUAAAUGAUCUGCUCAAGUGCUUGGGCAGCAAAUUGCCAAAGGGUUCGUGAUUUGA (((------(.((....)).))))..((((((........)))))).......((((((((....))))))))(((((..(.((....)).)..))))). ( -27.50) >DroSim_CAF1 57158 94 + 1 CAG------AAGCCAAGGCAUCUGGAAUUUCCAAAGAAGAGGAAAUAAAUGAUCUGCUCAAGUGCUUGGGCAGCAAAUUGCCAAAGGGUUCGUGAUUAGA ..(------((.((..((((..((..((((((........)))))).......((((((((....))))))))))...))))...)).)))......... ( -27.30) >DroEre_CAF1 46370 89 + 1 CCG------AAUCCAAGACAUCCGUGCUUUCAAAAGAAGAGGAAAUAAAUGACCUGCUCAAAUGCCUGGGCAGCAAACUGCCAAAGGGUUCGUA---A-- .((------(((((......(((...((((.....)))).)))..........(((((((......)))))))............)))))))..---.-- ( -23.90) >DroYak_CAF1 45985 92 + 1 CGG------AAUCCAAGGCAUCCGUACUUUCAAAGGAAGAGGAAAUAAAUGACCUGCUCAAAUGCCUGGGCAGUAAAUUGCCAAAGGGUUCGUAAUAA-- ..(------(((((..(((((((...((((.....)))).)))..........(((((((......))))))).....))))...)))))).......-- ( -27.80) >DroPer_CAF1 50259 100 + 1 CAGGAACUCCUGCCAAGCAAUCGAAGGAUGUCAGGGGCUACGAAGUGAACGAGCUGCUGAAGUGCUUGGGCAGCAGAUUGCAGAUGGGUUAGAGACUGUU ((((....))))((((((((((....))).((((.((((.((.......)))))).))))..)))))))((((....)))).(((((........))))) ( -33.50) >consensus CAG______AAGCCAAGGCAUCUGGAAUUUCAAAAGAAGAGGAAAUAAAUGAUCUGCUCAAAUGCUUGGGCAGCAAAUUGCCAAAGGGUUCGUAAUUAGA ............((..(((((((....((((....)))).)))..........((((((((....)))))))).....))))...))............. (-12.96 = -13.77 + 0.81)



| Location | 15,079,461 – 15,079,551 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.53 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.911038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15079461 90 - 23771897 UCUAAUUA----CCCUUUGGCAGUUUGCUGCCCAAGCAUUUGAGUAGAUCAUUUAUUUCCUCUUCUUUUGAAAUUCCAGAUGCCUUGGCUU------CUG ((((.(((----..(((.(((((....))))).)))....))).))))..............(((....)))....((((.((....)).)------))) ( -18.80) >DroSec_CAF1 44481 94 - 1 UCAAAUCACGAACCCUUUGGCAAUUUGCUGCCCAAGCACUUGAGCAGAUCAUUUAUUUCCUCUUCUUUGGAAAUUCCAGAUGCCUUGGCUU------CUG .........(((.((...((((.((((((((.(((....))).)))).......((((((........))))))..))))))))..)).))------).. ( -24.30) >DroSim_CAF1 57158 94 - 1 UCUAAUCACGAACCCUUUGGCAAUUUGCUGCCCAAGCACUUGAGCAGAUCAUUUAUUUCCUCUUCUUUGGAAAUUCCAGAUGCCUUGGCUU------CUG .........(((.((...((((.((((((((.(((....))).)))).......((((((........))))))..))))))))..)).))------).. ( -24.30) >DroEre_CAF1 46370 89 - 1 --U---UACGAACCCUUUGGCAGUUUGCUGCCCAGGCAUUUGAGCAGGUCAUUUAUUUCCUCUUCUUUUGAAAGCACGGAUGUCUUGGAUU------CGG --.---..((((.((...(((((....))))).(((((((((.(((((..........))).(((....))).)).))))))))).)).))------)). ( -27.70) >DroYak_CAF1 45985 92 - 1 --UUAUUACGAACCCUUUGGCAAUUUACUGCCCAGGCAUUUGAGCAGGUCAUUUAUUUCCUCUUCCUUUGAAAGUACGGAUGCCUUGGAUU------CCG --.......(((.((...((((......)))).(((((((((.((...(((.................)))..)).))))))))).)).))------).. ( -21.03) >DroPer_CAF1 50259 100 - 1 AACAGUCUCUAACCCAUCUGCAAUCUGCUGCCCAAGCACUUCAGCAGCUCGUUCACUUCGUAGCCCCUGACAUCCUUCGAUUGCUUGGCAGGAGUUCCUG ..(((.((((...(((...((((((((((.....))))..((((..(((((.......)).)))..))))........)))))).)))..))))...))) ( -27.90) >consensus UCUAAUCACGAACCCUUUGGCAAUUUGCUGCCCAAGCACUUGAGCAGAUCAUUUAUUUCCUCUUCUUUGGAAAUUCCAGAUGCCUUGGCUU______CUG .............((...(((.(((((((((.(((....))).))))...............(((....)))....))))))))..))............ (-10.14 = -10.53 + 0.39)

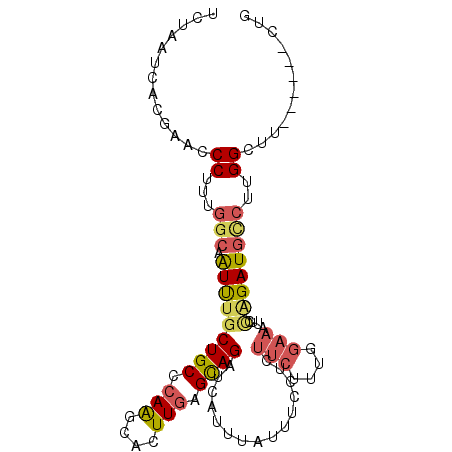
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:48 2006