
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,075,152 – 15,075,244 |
| Length | 92 |
| Max. P | 0.838330 |

| Location | 15,075,152 – 15,075,244 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -23.19 |
| Energy contribution | -24.97 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15075152 92 + 23771897 AAGGAGGCGGAGGACUUGAGCAGAAAGC---CACCUCCGCCACAGCAAAUGACAUCUCCAGUGGCACCCUCACUACAAGUGCUAGUGGAACCACC ..((.((((((((...((.((.....))---))))))))))...............((((.((((((...........)))))).)))).))... ( -35.30) >DroSec_CAF1 39813 92 + 1 AAGGAGGCGGAGGACUUGAGCAGAAAGC---CACCUCCGCCACAGCAACAGACAUCUCCAGUGGCACCCUCACUACAAGUGCUGGUGGAACCACC ..((.((((((((...((.((.....))---))))))))))...............((((.((((((...........)))))).)))).))... ( -34.00) >DroSim_CAF1 41113 92 + 1 AAGGAGGCGGAGGACUUGAGCAGAAAGC---CACCUCCGCCACAGCACCAGACAUCUCCAGUGGCACCCUCACUACAAGUGCUGGUGGAACCACC ..((.((((((((...((.((.....))---))))))))))....((((((.(((....(((((.....)))))....)))))))))...))... ( -36.70) >DroEre_CAF1 41014 95 + 1 AAGGAGGCGGAAGAUUUAAGCAGAAAGCCGCCACCUCCACCACCGCAACAGAUAUCUCCAGUAGCACCCUCACUUCAAGUGCUGGUGGAACCACC ..(((((.((....(((......)))....)).)))))..................((((.((((((...........)))))).))))...... ( -26.70) >DroYak_CAF1 41264 92 + 1 AAGGAGGCGGAAGAUUUGAGCAGAAAGC---CACCUCCACCACAGCAACAGAUAUCUCCAGUGGCACCCGCACUUCAAGUGCUGGUGGAACCACC ..(((((.((....(((......))).)---).)))))......................((((((((.((((.....)))).))))...)))). ( -28.40) >DroAna_CAF1 38514 83 + 1 AAGGAGGCCGAGGAUCUGAGCAAGAAGC---CCCCUCUGCAGCAGCAUCCGC---------AGGUUCCUCUGGUCCAAAUGCUGGUGGAGCCACC ..((.....(.(((((((.(((.((...---....)))))..))).)))).)---------.((((((.(..((......))..).)))))).)) ( -26.50) >consensus AAGGAGGCGGAGGACUUGAGCAGAAAGC___CACCUCCGCCACAGCAACAGACAUCUCCAGUGGCACCCUCACUACAAGUGCUGGUGGAACCACC ..((.((((((((......((.....)).....))))))))...............((((.((((((...........)))))).)))).))... (-23.19 = -24.97 + 1.78)



| Location | 15,075,152 – 15,075,244 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -26.49 |
| Energy contribution | -26.30 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15075152 92 - 23771897 GGUGGUUCCACUAGCACUUGUAGUGAGGGUGCCACUGGAGAUGUCAUUUGCUGUGGCGGAGGUG---GCUUUCUGCUCAAGUCCUCCGCCUCCUU (((((...((((..(((.....)))..)))))))))((((....(((.....)))(((((((.(---((.....))).....))))))))))).. ( -35.30) >DroSec_CAF1 39813 92 - 1 GGUGGUUCCACCAGCACUUGUAGUGAGGGUGCCACUGGAGAUGUCUGUUGCUGUGGCGGAGGUG---GCUUUCUGCUCAAGUCCUCCGCCUCCUU (((((...((((..(((.....)))..)))))))))((((((....))).....((((((((.(---((.....))).....))))))))))).. ( -35.70) >DroSim_CAF1 41113 92 - 1 GGUGGUUCCACCAGCACUUGUAGUGAGGGUGCCACUGGAGAUGUCUGGUGCUGUGGCGGAGGUG---GCUUUCUGCUCAAGUCCUCCGCCUCCUU ((((....)))).((((((.......))))))(((..((....))..)))....((((((((.(---((.....))).....))))))))..... ( -38.30) >DroEre_CAF1 41014 95 - 1 GGUGGUUCCACCAGCACUUGAAGUGAGGGUGCUACUGGAGAUAUCUGUUGCGGUGGUGGAGGUGGCGGCUUUCUGCUUAAAUCUUCCGCCUCCUU ((((.(((((..(((((((.......)))))))..))))).))))......((.((((((((.(((((....))))).....)))))))).)).. ( -37.90) >DroYak_CAF1 41264 92 - 1 GGUGGUUCCACCAGCACUUGAAGUGCGGGUGCCACUGGAGAUAUCUGUUGCUGUGGUGGAGGUG---GCUUUCUGCUCAAAUCUUCCGCCUCCUU (((((...((((.((((.....)))).)))))))))(((.(((.(....).)))((((((((.(---((.....))).....))))))))))).. ( -33.40) >DroAna_CAF1 38514 83 - 1 GGUGGCUCCACCAGCAUUUGGACCAGAGGAACCU---------GCGGAUGCUGCUGCAGAGGGG---GCUUCUUGCUCAGAUCCUCGGCCUCCUU ((((....)))).......((.((.(((((..((---------((((......))))))...((---((.....))))...)))))))))..... ( -32.30) >consensus GGUGGUUCCACCAGCACUUGUAGUGAGGGUGCCACUGGAGAUGUCUGUUGCUGUGGCGGAGGUG___GCUUUCUGCUCAAAUCCUCCGCCUCCUU ((((....)))).((((((.......))))))......................((((((((.....((.....))......))))))))..... (-26.49 = -26.30 + -0.19)


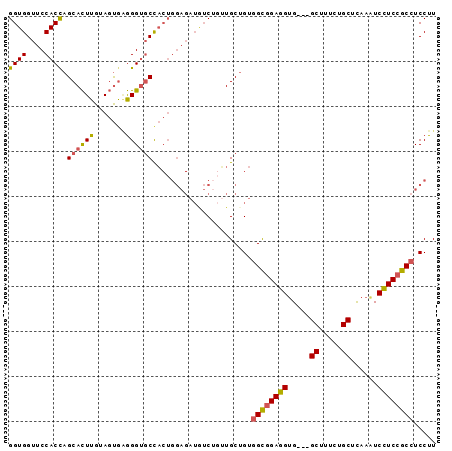
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:45 2006