
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,047,998 – 15,048,223 |
| Length | 225 |
| Max. P | 0.708032 |

| Location | 15,047,998 – 15,048,118 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15047998 120 - 23771897 AUGAGUCUAAAACUCUGCUGCAGCCAGGAGCACAUAAAACUGGGGGCAGCAGUUCCAGUUUGGUGAGUUUAAAUAUGCUUAAGCAGCUGGAGGCCAAGGUACAGGAACUACACACCCAAC .((.(((.......(((((((..((((............))))..))))))).(((((((...(((((........)))))...)))))))))))).(((...(......)..))).... ( -35.90) >DroPse_CAF1 15934 120 - 1 ACGAGUCAAAAACACUGCUGCAGCAGGCGGCCAAGGCAUCGCAAGGCAGCCAUUCCAGCUUGGUGAGCUUGAACAUGCUCAGACAGCUGGAGGCCAAGGUCCAGGAGCUGCACACCCAGC ..(..((.......((((....))))..((((..(((.((....))..)))..(((((((.(.(((((........)))))..))))))))))))..))..)....((((......)))) ( -45.10) >DroSim_CAF1 15056 120 - 1 AUGAGUCCAAAACUCUGCUGCAGCCAGGAGCACAUAAAACUGGAGGCAGCAGUUCCAGUUUGGUGAGUUUAAAUAUGCUAAAGCAACUGGAGGCCAAGGUACAGGAGCUACACACCCAAC .........((((((((((((..((((............))))..)))))))....)))))(((((((........)))..(((..(((...((....)).)))..)))...)))).... ( -36.50) >DroEre_CAF1 15497 120 - 1 ACGAGUCUAAAACUCUGCUGCAGCCAGGAGCACAUAAAGCUGGAGGCAGCAGUUCCAGUUUGGUGAGUCUAAACAUGCUUAAGCAACUGGAGGCCAAAGUGCAGGAACUGCACACCCAAC ..(.(((.......(((((((..((((............))))..))))))).(((((((...(((((........)))))...)))))))))))...((((((...))))))....... ( -41.70) >DroAna_CAF1 15499 120 - 1 AUGAGUCCAAAACGCUGCUCCAGGCAGGGGCAACUAAGGUUGGAGGCAGCAGUUCCAGUUUGGCCAGCUUAAACAUGCUGAGGCAACUGGAGGCCAAGGUCCAGGAACUGCACACCCAGC .............((((((((((.(..((....))..).))))))...(((((((((.((((((((((........))).((....))...))))))).)...)))))))).....)))) ( -47.70) >DroPer_CAF1 15957 120 - 1 ACGAGUCAAAAACACUGCUGCAGCAGGCGGCCAAGGCAUCGCAAGGCAGCCAUUCCAGCUUGGUGAGCUUGAACAUGCUCAGGCAGCUGGAGGCCAAGGUCCAGGAGCUGCACACCCAGC ..(..((.......((((....))))..((((..(((.((....))..)))..(((((((...(((((........)))))...)))))))))))..))..)....((((......)))) ( -45.00) >consensus ACGAGUCAAAAACUCUGCUGCAGCCAGGAGCAAAUAAAACUGGAGGCAGCAGUUCCAGUUUGGUGAGCUUAAACAUGCUCAAGCAACUGGAGGCCAAGGUCCAGGAACUGCACACCCAAC ..(.(((.......(((((((...(((............)))...))))))).(((((((...(((((........)))))...)))))))))))..(((......)))........... (-25.83 = -26.08 + 0.25)


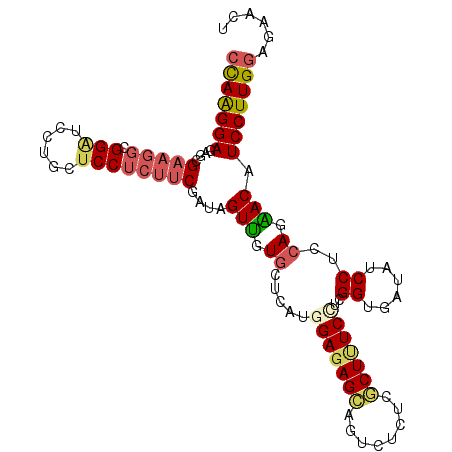
| Location | 15,048,118 – 15,048,223 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -26.15 |
| Energy contribution | -26.57 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15048118 105 + 23771897 CCAAGGAGAGGGAAGGCGGAUCCUGCUCCUCUUCCAUAGUUGUGCUCAUCGAGAGAAGUCUCUCGCUUUCUUCGGUAAUAUCCUCCAGAACAUCCUUGGAGAACU (((((((...((((((.(((......)))))))))...(((.((.....((((((....))))))........((......))..)).))).)))))))...... ( -34.90) >DroGri_CAF1 15983 96 + 1 CCAGGGACAGCGAUUUCGGC------UCCU---CCACUGUGGUGCUCAGCGAAAGUAGACGUUCACUCUCCUCGGUAAUGUCCUCGAGCACAUCCUUGCUGAACU ...((..((((((....((.------....---))......((((((.((....)).((((((.(((......)))))))))...))))))....))))))..)) ( -28.20) >DroSim_CAF1 15176 105 + 1 CCAAGGAGAGGGAAGGCGGAUCCUGCUCCUCUUCGAUAGUUGUGCUCAUGGAGAGCAGUCUCUCGCUUUCUUCGGUUAUAUCCUCCAGAACAUCCUUGGAGAACU (((((((((((.((((((((..((((((.(((..((.........))..)))))))))..)).))))))))))((......)).........)))))))...... ( -38.10) >DroEre_CAF1 15617 105 + 1 CUAAGGAGAGGGAAGGCGGAUCCUGCUCCUCUUCGACAGUUGUGCUCAGGGAGAGCAUUCUCUCGCUUUCCUCGGUGAUAUCCUCCAGAACAUCCUUGGAGAACU (((((((((((.((((((((...(((((.(((..((((....)).))..))))))))...)).))))))))))((.(.....).))......)))))))...... ( -37.30) >DroYak_CAF1 15682 105 + 1 CCAAGGAGAGCGAAGGCGGAUCCUGCUCCUCUUCGAUAGUUGUGCUCAUGGAGAGCAGUCUCUCGCUUUCCUCGGUGAUAUCCUCCAGGACAUCCUUGGAGAACU ((((((((((.(((((((((..((((((.(((..((.........))..)))))))))..)).)))))))))).......(((....)))..)))))))...... ( -42.00) >DroAna_CAF1 15619 105 + 1 CCAGGGACAGCGAAGGUGGGUCCUGCUCCUCUUCAAUGGUGGUGCUCAUGGAGAGCUGUCGUUCGCUCUCCUCGGUGAUAUCCUCCAGCACGUCCUUGGAGAACU ((((((((.(((((((.(((.....))).)))))..(((.((...(((((((((((........)))))))...))))...)).)))))..))))))))...... ( -44.90) >consensus CCAAGGAGAGCGAAGGCGGAUCCUGCUCCUCUUCGAUAGUUGUGCUCAUGGAGAGCAGUCUCUCGCUUUCCUCGGUGAUAUCCUCCAGAACAUCCUUGGAGAACU (((((((....(((((.(((......))))))))....(((.((.....(((((((........)))))))..((......))..)).))).)))))))...... (-26.15 = -26.57 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:37 2006