
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,978,005 – 14,978,115 |
| Length | 110 |
| Max. P | 0.998992 |

| Location | 14,978,005 – 14,978,115 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14978005 110 + 23771897 AUGUCUGUCUGU--UCAACAAUUUAAAUGUAUUUAACAUUAAAUUUUGAAACCAUUUAGAUAUUUUUUGCACUGCCUGUCUGUGGCAGUGUCAAAGCGCAUACCAAAGCAGG (((((((..(((--((((.(((((((.(((.....)))))))))))))))..))..))))))).((((((((((((.......))))))).))))).((........))... ( -28.80) >DroSec_CAF1 30163 89 + 1 AUGUCUGUUUUUUUUUUAACAUUAAAAUGU-----------------------AUUCAGAUAUUUUUUGCACUGCCUGUCUGUGGCAGUGUCAAAGCGCAUACCAAAACAGG (((((((..(...(((((....)))))...-----------------------)..))))))).((((((((((((.......))))))).)))))................ ( -22.00) >DroSim_CAF1 31160 88 + 1 AUGUCUGUUUUU-UUCAAAAAUUAAAAUGU-----------------------AUUCAGAUAUUUUUUGCACUGCCUGUCUGUGGCAGUGUCAAAGCGCAUACCAAAACAGG ....(((((((.-...........((((((-----------------------......))))))(((((((((((.......))))))).)))).........))))))). ( -21.50) >DroEre_CAF1 30215 108 + 1 AUUUCUGACUUU--UUAACUAUUA-AAUGUGUUCAACAUACAA-UCAGAAACCAUUCAGAUAUUUUUUGCACUGCCUGUCUGUGGCAGUGUCAAAGCGCAUACCAAAGCAGG .(((((((...(--(((.....))-))(((((.....))))).-)))))))((...........((((((((((((.......))))))).))))).((........)).)) ( -24.70) >DroYak_CAF1 31916 109 + 1 AUGUCUGACUGU--UCAGCUAUUAAAAUGUAUGCAACAUGCAAGUGAGAAACCAUUCACAUAUUUCUUGCACUGCC-GUCUGUGGCAGUGUCAAAGCGCAUACCAAAGCAGG ........((((--(..(((((......))).))...((((..(((((......))))).......((((((((((-(....)))))))).)))...)))).....))))). ( -28.60) >consensus AUGUCUGUCUUU__UCAACAAUUAAAAUGU_U__AACAU__AA_U__GAAACCAUUCAGAUAUUUUUUGCACUGCCUGUCUGUGGCAGUGUCAAAGCGCAUACCAAAGCAGG (((((((.................................................))))))).((((((((((((.......))))))).)))))................ (-16.01 = -16.45 + 0.44)

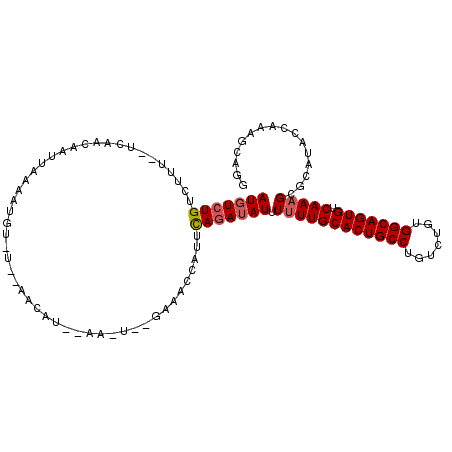
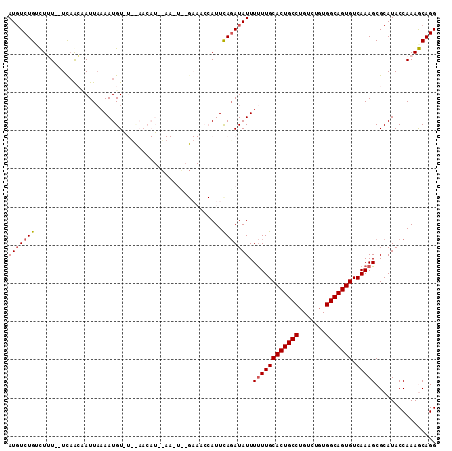
| Location | 14,978,005 – 14,978,115 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14978005 110 - 23771897 CCUGCUUUGGUAUGCGCUUUGACACUGCCACAGACAGGCAGUGCAAAAAAUAUCUAAAUGGUUUCAAAAUUUAAUGUUAAAUACAUUUAAAUUGUUGA--ACAGACAGACAU .(((((..(((((....((((.(((((((.......)))))))))))..)))))......((((...((((((((((.....))).)))))))...))--)))).))).... ( -26.70) >DroSec_CAF1 30163 89 - 1 CCUGUUUUGGUAUGCGCUUUGACACUGCCACAGACAGGCAGUGCAAAAAAUAUCUGAAU-----------------------ACAUUUUAAUGUUAAAAAAAAAACAGACAU .((((((((((((....((((.(((((((.......)))))))))))..))))).....-----------------------((((....)))).......))))))).... ( -23.80) >DroSim_CAF1 31160 88 - 1 CCUGUUUUGGUAUGCGCUUUGACACUGCCACAGACAGGCAGUGCAAAAAAUAUCUGAAU-----------------------ACAUUUUAAUUUUUGAA-AAAAACAGACAU .((((((((((((....((((.(((((((.......)))))))))))..))))).....-----------------------...(((((.....))))-)))))))).... ( -23.30) >DroEre_CAF1 30215 108 - 1 CCUGCUUUGGUAUGCGCUUUGACACUGCCACAGACAGGCAGUGCAAAAAAUAUCUGAAUGGUUUCUGA-UUGUAUGUUGAACACAUU-UAAUAGUUAA--AAAGUCAGAAAU ((......(((((....((((.(((((((.......)))))))))))..))))).....))(((((((-((...(((((((....))-))))).....--..))))))))). ( -28.90) >DroYak_CAF1 31916 109 - 1 CCUGCUUUGGUAUGCGCUUUGACACUGCCACAGAC-GGCAGUGCAAGAAAUAUGUGAAUGGUUUCUCACUUGCAUGUUGCAUACAUUUUAAUAGCUGA--ACAGUCAGACAU .........((((((..((((.(((((((......-))))))))))).((((((..(.(((....))).)..)))))))))))).........(((((--....)))).).. ( -34.10) >consensus CCUGCUUUGGUAUGCGCUUUGACACUGCCACAGACAGGCAGUGCAAAAAAUAUCUGAAUGGUUUC__A_UU__AUGUU__A_ACAUUUUAAUAGUUGA__AAAGACAGACAU .((((((((((((....((((.(((((((.......)))))))))))..))))).(((((.......................)))))............)))).))).... (-17.44 = -17.88 + 0.44)

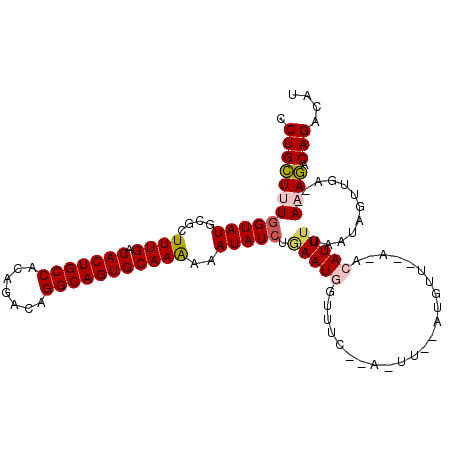
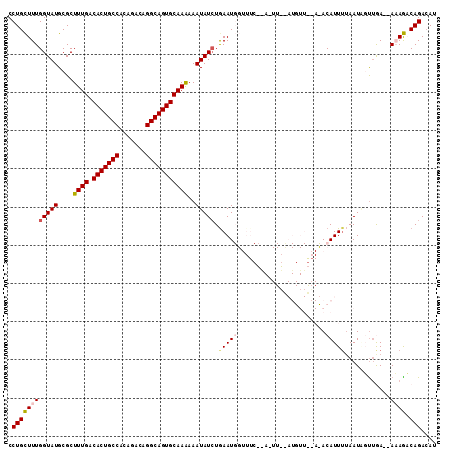
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:21 2006