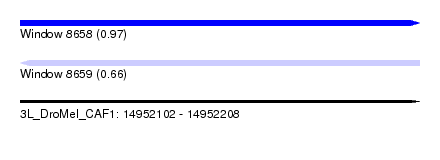
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,952,102 – 14,952,208 |
| Length | 106 |
| Max. P | 0.973171 |

| Location | 14,952,102 – 14,952,208 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.14 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14952102 106 + 23771897 ACAUUUAACCUUGUUACAAUUAUUGUGCACACAAAUAGUUUGUGGCAAGUUUCACUUGCAGUAU--------------UUGUCCCACAAAGCCGUUAAAUCAGGUGCAAUUAAAGGCUUA ...........((..((((.((((((((.(((((.....)))))))(((.....))))))))).--------------))))..))..(((((.((((...........)))).))))). ( -21.70) >DroSec_CAF1 3885 90 + 1 ACAUUUAACCUUGUUACAAUUAUUGUGCACACAAAUAGUUUGUGGCAAGUUUUACUUGCA------------------------------GCCGUUAAAUCAGCUGCAAUUAAAGUCCUA .........(((((((((((((((((....)).))))).))))))))))(((((.(((((------------------------------((.(......).))))))).)))))..... ( -24.80) >DroSim_CAF1 3931 90 + 1 ACAUUUAACCUUGUUACAAUUAUUGUGCGCACAAAUAGUUUGUGGCAAGUUUUACUUGCA------------------------------GCCGUUAAAUCAGCUGCAAUUAAAGGCAUA .........(((((((((((((((((....)).))))).))))))))))(((((.(((((------------------------------((.(......).))))))).)))))..... ( -25.00) >DroEre_CAF1 4040 120 + 1 ACGUUCAGCCUUGUCACAAUUAUUGUGCACACAAAUAGCUUGUGGCAAGUUUAACUUGCAGUACAAGUAUGUACAUAUAUGUCCCACGAAGCCGUUAAAUCAGCUACAAUUAAAGGCCUA .......(((((((((((.....)))).)).........(((((((..(((((((..((.(((((....))))).....((.....))..)).)))))))..)))))))...)))))... ( -29.30) >DroYak_CAF1 4231 106 + 1 ACGUUCAACCUUGUUACAAUUGUUGUGCACACAAAUAGUUUGUGGCAAGUUUCACUUGCAGUAUGUGUA---------AUGUCCCA-----CCGUUAAAUCAGCUGCAAUUACAGGCCUA ..(..(...((((((((((...((((....)))).....))))))))))......(((((((.((..((---------(((.....-----.)))))...))))))))).....)..).. ( -25.00) >consensus ACAUUUAACCUUGUUACAAUUAUUGUGCACACAAAUAGUUUGUGGCAAGUUUCACUUGCAGUA________________UGUCCCA____GCCGUUAAAUCAGCUGCAAUUAAAGGCCUA ........((((.........(((((((.(((((.....)))))(((((.....)))))...........................................)).)))))..)))).... (-14.86 = -15.14 + 0.28)


| Location | 14,952,102 – 14,952,208 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -24.74 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14952102 106 - 23771897 UAAGCCUUUAAUUGCACCUGAUUUAACGGCUUUGUGGGACAA--------------AUACUGCAAGUGAAACUUGCCACAAACUAUUUGUGUGCACAAUAAUUGUAACAAGGUUAAAUGU ..((((((...(((((.(((......)))..(((((.(((((--------------(((..(((((.....))))).......))))))).).)))))....))))).))))))...... ( -21.60) >DroSec_CAF1 3885 90 - 1 UAGGACUUUAAUUGCAGCUGAUUUAACGGC------------------------------UGCAAGUAAAACUUGCCACAAACUAUUUGUGUGCACAAUAAUUGUAACAAGGUUAAAUGU ...((((((..(((((((((......))))------------------------------)))))........((((((((.....))))).))).............))))))...... ( -24.60) >DroSim_CAF1 3931 90 - 1 UAUGCCUUUAAUUGCAGCUGAUUUAACGGC------------------------------UGCAAGUAAAACUUGCCACAAACUAUUUGUGCGCACAAUAAUUGUAACAAGGUUAAAUGU ...(((((...(((((((((......))))------------------------------)))))........((((((((.....))))).))).............)))))....... ( -24.90) >DroEre_CAF1 4040 120 - 1 UAGGCCUUUAAUUGUAGCUGAUUUAACGGCUUCGUGGGACAUAUAUGUACAUACUUGUACUGCAAGUUAAACUUGCCACAAGCUAUUUGUGUGCACAAUAAUUGUGACAAGGCUGAACGU ..((((((((((((((((((......)))))..............(((((((((((((...(((((.....))))).))))).....))))))))..)))))))....))))))...... ( -31.20) >DroYak_CAF1 4231 106 - 1 UAGGCCUGUAAUUGCAGCUGAUUUAACGG-----UGGGACAU---------UACACAUACUGCAAGUGAAACUUGCCACAAACUAUUUGUGUGCACAACAAUUGUAACAAGGUUGAACGU ..(((.(((....))))))..((((((..-----((..(((.---------..........(((((.....)))))(((((.....)))))...........)))..))..))))))... ( -21.40) >consensus UAGGCCUUUAAUUGCAGCUGAUUUAACGGC____UGGGACA________________UACUGCAAGUAAAACUUGCCACAAACUAUUUGUGUGCACAAUAAUUGUAACAAGGUUAAAUGU ..((((((...(((((((((......))))...............................(((((.....)))))((((((...))))))...........))))).))))))...... (-14.88 = -15.12 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:14 2006