
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,944,626 – 14,944,903 |
| Length | 277 |
| Max. P | 0.977153 |

| Location | 14,944,626 – 14,944,746 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -31.08 |
| Energy contribution | -31.76 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14944626 120 + 23771897 UCAUUUCCUUUUCGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGA ((((((((.((((.((((((........(((.......))).........))))))...(((((((....(.(((((.......))))).)..)))))))...))))....)))))).)) ( -34.13) >DroSec_CAF1 125002 120 + 1 UCAUUUCCUUUUUGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAGGCGUGCGCUCCUCAAGCGCAGGACACGUACGGAAGAAACCGUGGUAUUCGA ....((((...((.((((((........(((.......))).........)))))).)).((((((....(.((((((.....)))))).)..))))))))))(((((....)).))).. ( -33.03) >DroSim_CAF1 122710 120 + 1 UCAUUUCCUUUUUGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGA ((((((((.(((((((((((........(((....))).................(((.((((((.....)))))).)))))))).)))))).....((((......)))))))))).)) ( -35.50) >DroEre_CAF1 119731 117 + 1 UCAUUUCCUUUCUGCCGUGACUCCUUUCGAGAACCCU---CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAAUCGUGGGAAUCGA ((..((((.(((((((((((........(((....))---)..............(((.((((((.....)))))).)))))))).))))))((((..(....)....))))))))..)) ( -38.70) >DroYak_CAF1 120720 117 + 1 UCAUUUCCUUUUUUCCGUGACUCCUUUCGAGAACCCU---CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGA ((((((((...(((((((((........(((....))---).........)))))))))(((((((....(.(((((.......))))).)..)))))))...........)))))).)) ( -35.23) >consensus UCAUUUCCUUUUUGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGA ((((((((.((((((((((((((.....)))........................(((.((((((.....)))))).)))))))).)))))).....((((......)))))))))).)) (-31.08 = -31.76 + 0.68)

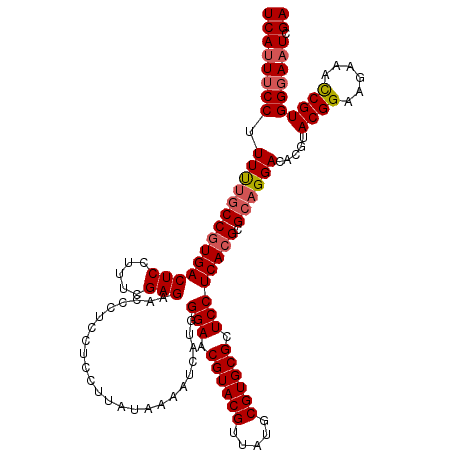

| Location | 14,944,666 – 14,944,786 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -28.12 |
| Energy contribution | -27.64 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14944666 120 + 23771897 CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGAAACCGAAAAUGCACCUAAACAAAAAUAAAUGCACCAGACC ...........(((((...(((((((....(.(((((.......))))).)..))))))).......)))))((..(((....)))...((((..((........))..))))))..... ( -30.00) >DroSec_CAF1 125042 120 + 1 CCUUAUAAAAUCAUGGGAACGUACGUUAGGCGUGCGCUCCUCAAGCGCAGGACACGUACGGAAGAAACCGUGGUAUUCGAAACCGAAAAUGCGCCUGAACAAAGAUAAAUGCACCAGACC .........(((...(((.((((((.....)))))).)))(((.(((((....((.(((((......))))))).((((....))))..))))).))).....))).............. ( -33.10) >DroSim_CAF1 122750 120 + 1 CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGAAACCGAAAAUGCACCUAAACAAAAAUAAAUGCACCAGACC ...........(((((...(((((((....(.(((((.......))))).)..))))))).......)))))((..(((....)))...((((..((........))..))))))..... ( -30.00) >DroEre_CAF1 119768 119 + 1 CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAAUCGUGGGAAUCGAAACCGA-AAUGCACCUAAACAAAAAUAAAUGCAGUAGACC ..........((((((...(((((((....(.(((((.......))))).)..))))))).......))))))...(((....)))-..((((..((........))..))))....... ( -26.50) >DroYak_CAF1 120757 119 + 1 CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGAAACCGA-AAUGCACCUAAACAAAAAUAAAUGCACCAGACC ...........(((((...(((((((....(.(((((.......))))).)..))))))).......)))))((..(((....)))-..((((..((........))..))))))..... ( -30.00) >consensus CCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCGAAACCGAAAAUGCACCUAAACAAAAAUAAAUGCACCAGACC ..........((((((...(((((((....(.(((((.......))))).)..))))))).......))))))...(((....)))...((((..((........))..))))....... (-28.12 = -27.64 + -0.48)



| Location | 14,944,746 – 14,944,864 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -35.56 |
| Consensus MFE | -31.16 |
| Energy contribution | -31.64 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14944746 118 - 23771897 CCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA--GGGACCUGUCUAUUGUUACCCUCUUCUAGGGUCUGGUGCAUUUAUUUUUGUUUAGGUGCAUUUUCGGUU .(((((((((.....((((...........))))..)))))))))((((--(((((((.......(.....).......)))))).(((((((((........))))))))))))))... ( -36.04) >DroSec_CAF1 125122 115 - 1 CCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA--GGGACCUGUCUAUG--U-ACCUCUUCUAAGGUCUGGUGCAUUUAUCUUUGUUCAGGCGCAUUUUCGGUU ((((((((((.....((((...........))))..))))))))(((((--((.((........)--)-.)))))......((((((.(((........)))))))))))......)).. ( -34.00) >DroSim_CAF1 122830 118 - 1 CCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA--GGGACCUGUCUAUUGUUACCCUCUUCUAGGGUCUGGUGCAUUUAUUUUUGUUUAGGUGCAUUUUCGGUU .(((((((((.....((((...........))))..)))))))))((((--(((((((.......(.....).......)))))).(((((((((........))))))))))))))... ( -36.04) >DroEre_CAF1 119848 117 - 1 CCACCUGGGCAACCUUUUCUAAUUCAAUUCGAAAUGGCCCGGCUGCGGA--GGGACCUGUCUAUUGUUACCCUCUUCUAGGGUCUACUGCAUUUAUUUUUGUUUAGGUGCAUU-UCGGUU ..(((.((....))................((((((((((....(.(((--(((...............)))))).)..)))))....(((((((........))))))))))-))))). ( -32.66) >DroYak_CAF1 120837 119 - 1 CCACCUGGGCAACCUUUUCUAAUUCAAUUCGAAAUGGCCCGGGUGCGGAGAGGGACCUGUCUAUUGUUACCCUCUUCUAGGGUCUGGUGCAUUUAUUUUUGUUUAGGUGCAUU-UCGGUU .(((((((((.....((((...........))))..))))))))).((((((((...............))))))))......((((((((((((........))))))))).-.))).. ( -39.06) >consensus CCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA__GGGACCUGUCUAUUGUUACCCUCUUCUAGGGUCUGGUGCAUUUAUUUUUGUUUAGGUGCAUUUUCGGUU .(((((((((.....((((...........))))..)))))))))(((....((((((.....................))))))((((((((((........)))))))))).)))... (-31.16 = -31.64 + 0.48)

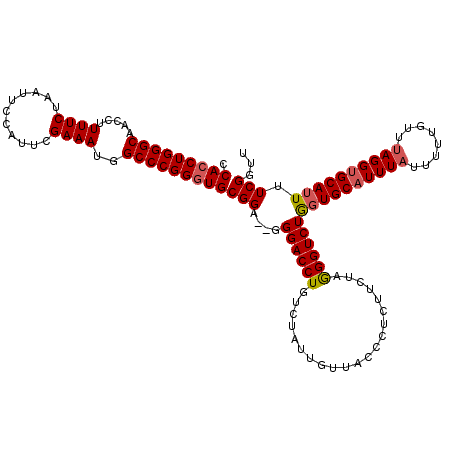

| Location | 14,944,786 – 14,944,903 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14944786 117 - 23771897 CUUUCAUUCUUCGG-GACCCAUUAAAAUCGACCCCCAACUCCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA--GGGACCUGUCUAUUGUUACCCUCUUCUAG ....((.....(((-(.(((.........(.....)..((.(((((((((.....((((...........))))..))))))))).)).--))).)))).....)).............. ( -28.90) >DroSec_CAF1 125162 115 - 1 CUUUCAUUCUUCGGGGGCCCAUUAAAAUCGACCCCCAACUCCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA--GGGACCUGUCUAUG--U-ACCUCUUCUAA ............(((((.(.((....)).).))))).....(((((((((.....((((...........))))..))))))))).(((--((.((........)--)-.)))))..... ( -33.80) >DroSim_CAF1 122870 117 - 1 CUUUCAUUCUUCGG-GGCCCAUUAAAAUCGACCCCCAACUCCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA--GGGACCUGUCUAUUGUUACCCUCUUCUAG ............((-((..(.........)..)))).....(((((((((.....((((...........))))..))))))))).(((--(((...............))))))..... ( -31.56) >DroEre_CAF1 119887 117 - 1 CUUUCAUUCUGCGG-GACCCAUUAAGAUCUACCCCCAACUCCACCUGGGCAACCUUUUCUAAUUCAAUUCGAAAUGGCCCGGCUGCGGA--GGGACCUGUCUAUUGUUACCCUCUUCUAG ............((-(...((...((((...(((((.....((.((((((.....((((...........))))..)))))).)).)).--)))....))))..))...)))........ ( -24.00) >DroYak_CAF1 120876 119 - 1 CUUUCAUUCUUCGG-GACCCAUUACAAUCUACCCCCAACUCCACCUGGGCAACCUUUUCUAAUUCAAUUCGAAAUGGCCCGGGUGCGGAGAGGGACCUGUCUAUUGUUACCCUCUUCUAG ............((-(.................))).....(((((((((.....((((...........))))..))))))))).((((((((...............))))))))... ( -32.89) >consensus CUUUCAUUCUUCGG_GACCCAUUAAAAUCGACCCCCAACUCCACCUGGGCAACCUUUUCUAAUUCCAUUCGAAAUGGCCCGGGUGCGGA__GGGACCUGUCUAUUGUUACCCUCUUCUAG ............((....))............(((...((.(((((((((.....((((...........))))..))))))))).))...))).......................... (-23.48 = -23.68 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:09 2006