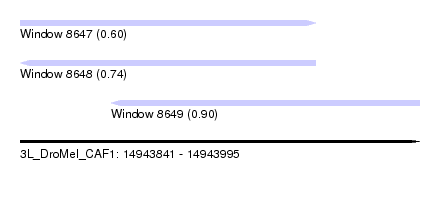
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,943,841 – 14,943,995 |
| Length | 154 |
| Max. P | 0.898820 |

| Location | 14,943,841 – 14,943,955 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600751 |
| Prediction | RNA |
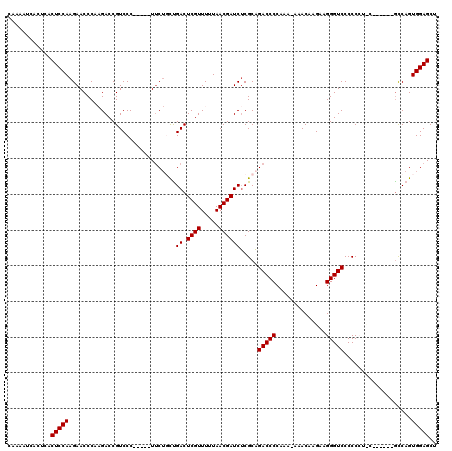
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14943841 114 + 23771897 CAAAAUCACUCACUCCAAGAACCCAAGACCGUCCC-----UUCUGCUGACUCGUUUUUAACGAUCUCGCAGACCCCAAA-AAACAAGAAGGGUCCCCCCUCCAACCCCACCAGUGGAGCU ............(((((..................-----.(((((.((.((((.....))))))..))))).......-......(.((((....)))).)...........))))).. ( -21.50) >DroSec_CAF1 124213 108 + 1 CAAAAUCACUCACUCCAAGAACCCAAGACCGUCCC-----UUCUGCUGACUCGUUUUUAACGAUCUCGCAGACCCCAAA-AAACAAGAAGGGUCCCCCCUUC------GUCAGUGGAGCU ............(((((.(....)..(((......-----.(((((.((.((((.....))))))..))))).......-......((((((....))))))------)))..))))).. ( -26.70) >DroSim_CAF1 121980 109 + 1 CAAAAUCACUCACUCCAAGAACCCAAGACCGUCCC-----UACUGCUGACUCGUUUUUAACGAUCUCGCAGACCCCAAAAAAACAAGAAGGGUCCCCCCUUC------GCCAGUGGAGCU ............(((((..................-----..((((.((.((((.....))))))..))))...............((((((....))))))------.....))))).. ( -24.90) >DroEre_CAF1 119082 96 + 1 CAAAGUCA----CUCCAAGAACCC--GACCGUCCC-----UUC-UUCGACUCGUUUUUAACGAUCUCGCAGACCCCAA--AAACAAGAAGGGUCCCCC----------GCCAGUGGAGCU ........----(((((......(--(...(.(((-----(((-(((((.((((.....))))..)))..........--....)))))))).)...)----------)....))))).. ( -21.13) >DroYak_CAF1 120044 102 + 1 CAAAAUCA----CUCCAAGAACCC--GACCGUCCCGUCCCUUCUUUUGACUCGUUUUUAACGAUCGCGCCGACCCCAA--AAACAAGAAGGGUCCCCC----------GCCAGUGGAGCU ........----(((((((((...--(((......)))..))))...((.((((.....))))))(((..(((((...--.........)))))...)----------))...))))).. ( -21.90) >consensus CAAAAUCACUCACUCCAAGAACCCAAGACCGUCCC_____UUCUGCUGACUCGUUUUUAACGAUCUCGCAGACCCCAAA_AAACAAGAAGGGUCCCCCCU_C______GCCAGUGGAGCU ............(((((..............................((.((((.....)))))).....(((((..............)))))...................))))).. (-15.62 = -15.62 + 0.00)

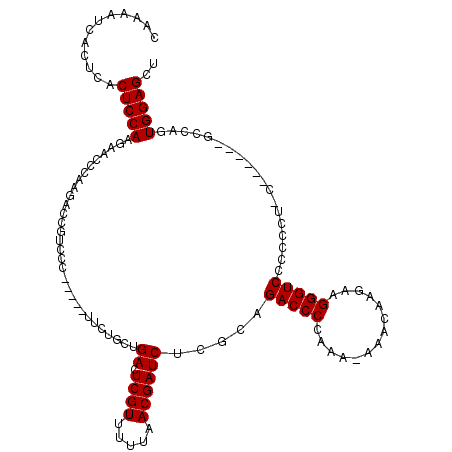
| Location | 14,943,841 – 14,943,955 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -26.28 |
| Energy contribution | -27.80 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14943841 114 - 23771897 AGCUCCACUGGUGGGGUUGGAGGGGGGACCCUUCUUGUUU-UUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGAA-----GGGACGGUCUUGGGUUCUUGGAGUGAGUGAUUUUG .(((((.(((.(((....((((((....))))))((((((-((.(.((((.....)))).).)))))))).))).))).(-----(((((........)))))))))))........... ( -40.50) >DroSec_CAF1 124213 108 - 1 AGCUCCACUGAC------GAAGGGGGGACCCUUCUUGUUU-UUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGAA-----GGGACGGUCUUGGGUUCUUGGAGUGAGUGAUUUUG .(((((.(((((------((((((....))))))((((((-((.(.((((.....)))).).)))))))))))))....(-----(((((........)))))))))))........... ( -42.00) >DroSim_CAF1 121980 109 - 1 AGCUCCACUGGC------GAAGGGGGGACCCUUCUUGUUUUUUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGUA-----GGGACGGUCUUGGGUUCUUGGAGUGAGUGAUUUUG .(((((.(((((------((((((....))))))((((((((..(.((((.....)))).).)))))))))))))....(-----(((((........)))))))))))........... ( -40.30) >DroEre_CAF1 119082 96 - 1 AGCUCCACUGGC----------GGGGGACCCUUCUUGUUU--UUGGGGUCUGCGAGAUCGUUAAAAACGAGUCGAA-GAA-----GGGACGGUC--GGGUUCUUGGAG----UGACUUUG .(((((((((((----------.(....((((((((((((--((((((((.....)))).))))))))......))-)))-----))).).)))--)).....)))))----)....... ( -32.20) >DroYak_CAF1 120044 102 - 1 AGCUCCACUGGC----------GGGGGACCCUUCUUGUUU--UUGGGGUCGGCGCGAUCGUUAAAAACGAGUCAAAAGAAGGGACGGGACGGUC--GGGUUCUUGGAG----UGAUUUUG .((((((.....----------..(((((((..(((((((--(((((((((...))))).)))))))))))...........(((......)))--))))))))))))----)....... ( -33.51) >consensus AGCUCCACUGGC______G_AGGGGGGACCCUUCUUGUUU_UUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGAA_____GGGACGGUCUUGGGUUCUUGGAGUGAGUGAUUUUG .(((((((((((......((((((....))))))((((((....(.(((((...))))).)...)))))))))))..........(((((........)))))))))))........... (-26.28 = -27.80 + 1.52)

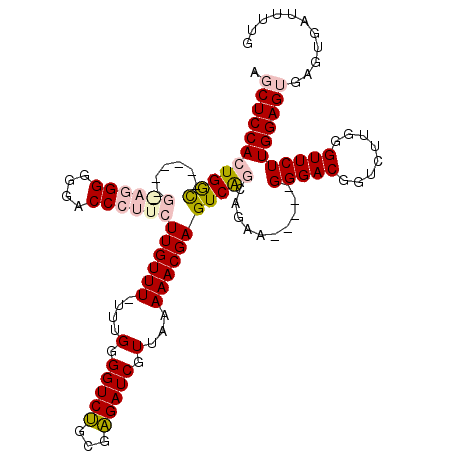
| Location | 14,943,876 – 14,943,995 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -25.81 |
| Energy contribution | -27.25 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
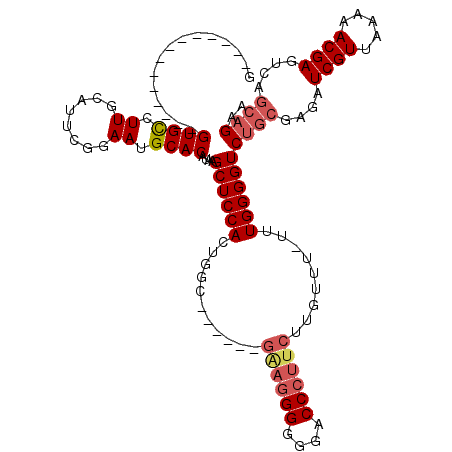
>3L_DroMel_CAF1 14943876 119 - 23771897 GGUGAGUAAGUGCCUGUGCCUUGCACUCGGAAUGCACAUAAGCUCCACUGGUGGGGUUGGAGGGGGGACCCUUCUUGUUU-UUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGAA .....(((((.((....))))))).(((((((.(((....(((((((....)))))))((((((....)))))).))).)-)))))).(((((....((((.....))))....))))). ( -46.90) >DroSec_CAF1 124248 99 - 1 G--------------GUGCCUUGCAUUCUGAAUGCACAUAAGCUCCACUGAC------GAAGGGGGGACCCUUCUUGUUU-UUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGAA (--------------(.((..(((((.....))))).....)).)).(((((------((((((....))))))((((((-((.(.((((.....)))).).)))))))))))))..... ( -37.60) >DroSim_CAF1 122015 100 - 1 G--------------GUGCCUUGCAUUCGGAAUGCACAUAAGCUCCACUGGC------GAAGGGGGGACCCUUCUUGUUUUUUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGUA (--------------(.((..(((((.....))))).....)).)).(((((------((((((....))))))((((((((..(.((((.....)))).).)))))))))))))..... ( -36.00) >DroYak_CAF1 120078 107 - 1 -GUGAGCGAGUGCCUGUGUCUUAUAUUCAGAAUGCACAUAAGCUCCACUGGC----------GGGGGACCCUUCUUGUUU--UUGGGGUCGGCGCGAUCGUUAAAAACGAGUCAAAAGAA -((.(((((((((((((((.((........)).)))))...((((((..(((----------(((((....)))))))).--.)))))).)))))..)))))....))............ ( -30.50) >consensus G______________GUGCCUUGCAUUCGGAAUGCACAUAAGCUCCACUGGC______GAAGGGGGGACCCUUCUUGUUU_UUUGGGGUCUGCGAGAUCGUUAAAAACGAGUCAGCAGAA ...............((((.((........)).))))....((((((...........((((((....)))))).........))))))((((....((((.....))))....)))).. (-25.81 = -27.25 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:05 2006