
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,942,639 – 14,942,904 |
| Length | 265 |
| Max. P | 0.888328 |

| Location | 14,942,639 – 14,942,749 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.76 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -21.73 |
| Energy contribution | -21.49 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14942639 110 - 23771897 CACCCUCUACGUAAUUCUACGUAACUUUGGAUGGUACCGAAGCUUUAAAGACCGCUUGGCUCUC------CCAUUUUCCAACGCCCACCAAUCCACC----CACCACUCAAGUGGAAGAA .......((((((....))))))....((((((((....((((..........))))(((....------............))).))).)))))..----..((((....))))..... ( -22.59) >DroSec_CAF1 122965 110 - 1 CACCCUCUACGUAGUUCUACGUAACUUUGGAUGGUACCGAAGCUUUAAAGACCGCUUGGCCCUC------CCAUUUCCCAACGCCCAUCAUUCCACC----CACCACUCAAGUGGGAGAA .......((((((....))))))....((((((((....((((..........))))(((....------............))).)))).))))((----(((.......))))).... ( -22.49) >DroSim_CAF1 120727 110 - 1 CACCCUCUACGUAAUUCUACGUAACUUUGGAUGGUACCGAAGCUUUAAAGACCGCUUGGCUCUC------CCAUUUCCCAACGCCCACCAUUCCACC----CACCACUCAAGUGGGAGAA .......((((((....))))))....((((((((....((((..........))))(((....------............))).)))).))))((----(((.......))))).... ( -24.89) >DroEre_CAF1 117730 109 - 1 CACCCACUACGUAGACCUACGUAACUUUGGAUGGUACCGAAGCUUUAAAGACCGCUUGGCUCCCCAUU-------CCCCAACGCCCACCAGUCCACC----CGCCACUCAAGUGGGAGAA ..(((((((((((....))))))....((((((((....((((..........))))(((........-------.......))).)))).))))..----..........))))).... ( -27.36) >DroYak_CAF1 118837 120 - 1 CACCCACUACGUAAUCCUACGUAACCUUGGAUGGUACCGAAGCUUUAAAGACCGCUUGGCUCCCCAUUACCCAUUACCCAACGCCCAUCAGUCCACCCACCCACCACUCAAGUGGGAGAG ..(((((((((((....))))))...(((..((((....((((..........))))(((......................))).................))))..)))))))).... ( -25.25) >consensus CACCCUCUACGUAAUUCUACGUAACUUUGGAUGGUACCGAAGCUUUAAAGACCGCUUGGCUCUC______CCAUUUCCCAACGCCCACCAGUCCACC____CACCACUCAAGUGGGAGAA .......((((((....))))))....((((((((....((((..........))))(((......................))).)))).))))........((((....))))..... (-21.73 = -21.49 + -0.24)

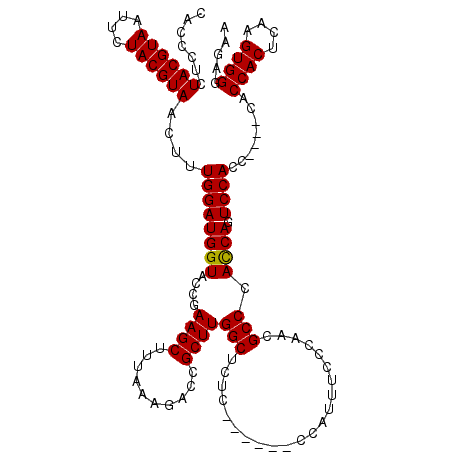

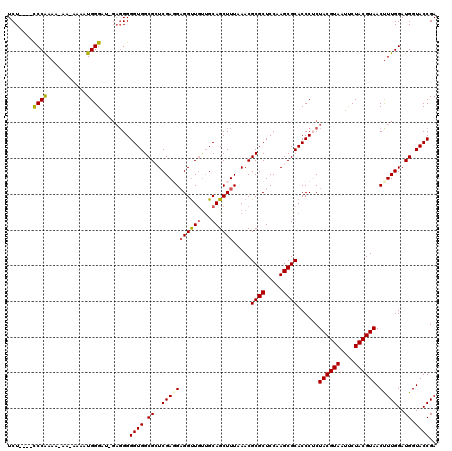
| Location | 14,942,709 – 14,942,824 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14942709 115 - 23771897 UCU----CCCAAAAAAAUUAAAUGGGAU-GAGGGGGUGGCGCUCGAGGUGGUUUUUGCAGCUUUAAACGCGCUCCAAGCGCACCCUCUACGUAAUUCUACGUAACUUUGGAUGGUACCGA (((----((((...........))))).-))(((((((.((((.(..(((((((..........)))).)))..).)))))))))))((((((....))))))...((((......)))) ( -37.50) >DroSec_CAF1 123035 114 - 1 UCU----CCCAAAAUAA-AAAAUGGGAU-GAGGGGGUGGCGCUCGACGAGGUUGUUGCAGCUUUAAACGCGCUCCAAGCGCACCCUCUACGUAGUUCUACGUAACUUUGGAUGGUACCGA (((----((((......-....))))).-))((((((...((((((((....))))).))).......((((.....))))))))))((((((....))))))...((((......)))) ( -36.30) >DroSim_CAF1 120797 114 - 1 UCU----CCCAAAAUAA-AAAAUGGGAU-GAGGGGGUGGCGCUCGACGAGGUUGUUGCAGCUUUAAACGCGCUCCAAGCGCACCCUCUACGUAAUUCUACGUAACUUUGGAUGGUACCGA (((----((((......-....))))).-))((((((...((((((((....))))).))).......((((.....))))))))))((((((....))))))...((((......)))) ( -35.60) >DroEre_CAF1 117799 108 - 1 UCU----CCCGAA-----GAAUUGGGAGC---GGGGUGGCGAUCGAGGAGGCUGCUGCAGCUUUAAACGCGCUCCAAGCGCACCCACUACGUAGACCUACGUAACUUUGGAUGGUACCGA ...----.(((((-----(...((((.((---(.((.((((......(((((((...))))))).....))))))...))).)))).((((((....)))))).)))))).......... ( -38.70) >DroYak_CAF1 118917 115 - 1 UCUCUCUUCCAAA-----GAAUUGGGAGAGAGGGGGUGGCGAUCGAGGAGGUUGUAGCAGCUUUAAACGCGCUCCAAGCGCACCCACUACGUAAUCCUACGUAACCUUGGAUGGUACCGA ((((((((((((.-----...)))))))))))).((((.((.((((((((((((...)))))).....((((.....))))......((((((....)))))).)))))).)).)))).. ( -46.00) >consensus UCU____CCCAAAA_AA_AAAAUGGGAU_GAGGGGGUGGCGCUCGAGGAGGUUGUUGCAGCUUUAAACGCGCUCCAAGCGCACCCUCUACGUAAUUCUACGUAACUUUGGAUGGUACCGA .......((((...........))))........((((.((..(((((((((((...)))))).....((((.....))))......((((((....)))))).)))))..)).)))).. (-29.20 = -29.44 + 0.24)


| Location | 14,942,749 – 14,942,864 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.53 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14942749 115 - 23771897 ACGAUCACAUCGCCUCAUCUCGUGAAACACCCACCCACUCUCU----CCCAAAAAAAUUAAAUGGGAU-GAGGGGGUGGCGCUCGAGGUGGUUUUUGCAGCUUUAAACGCGCUCCAAGCG ..........(((.((((((((.(......((((((.(.((((----((((...........))))).-))))))))))..).))))))))...(((.(((.........))).)))))) ( -36.70) >DroSec_CAF1 123075 114 - 1 ACGAUCACAUCGCCUCAUCUCGUGAAACACCCACCCACUCUCU----CCCAAAAUAA-AAAAUGGGAU-GAGGGGGUGGCGCUCGACGAGGUUGUUGCAGCUUUAAACGCGCUCCAAGCG ...........((..((((((((((.....((((((.(.((((----((((......-....))))).-))))))))))...)).)))))).))..)).........(((.......))) ( -35.70) >DroSim_CAF1 120837 114 - 1 ACGAUCACAUCGCCUCAUCUCGUAAAACACCCACCCACUCUCU----CCCAAAAUAA-AAAAUGGGAU-GAGGGGGUGGCGCUCGACGAGGUUGUUGCAGCUUUAAACGCGCUCCAAGCG ...........((..((((((((.......((((((.(.((((----((((......-....))))).-))))))))))......)))))).))..)).........(((.......))) ( -36.12) >DroEre_CAF1 117839 107 - 1 ACGAUCACAUCGCCUCAUCUCGUGCAACAC-CACCCACUCUCU----CCCGAA-----GAAUUGGGAGC---GGGGUGGCGAUCGAGGAGGCUGCUGCAGCUUUAAACGCGCUCCAAGCG ...........(((((..((((.....(.(-(((((.....((----(((((.-----...))))))).---.)))))).)..))))))))).(((..(((.........)))...))). ( -39.40) >DroYak_CAF1 118957 115 - 1 CCGAUCACAUCGCCUCAUCUCGUGAAAUACCCACCCACUCUCUCUCUUCCAAA-----GAAUUGGGAGAGAGGGGGUGGCGAUCGAGGAGGUUGUAGCAGCUUUAAACGCGCUCCAAGCG .(((((...((((........)))).........((((((((((((((((((.-----...)))))))))))))))))).)))))....(.(((.(((.((.......))))).))).). ( -47.50) >consensus ACGAUCACAUCGCCUCAUCUCGUGAAACACCCACCCACUCUCU____CCCAAAA_AA_AAAAUGGGAU_GAGGGGGUGGCGCUCGAGGAGGUUGUUGCAGCUUUAAACGCGCUCCAAGCG ...........(((((.((..(((.......)))(((((((((....((((...........))))....))))))))).....)).)))))...............(((.......))) (-26.42 = -26.74 + 0.32)

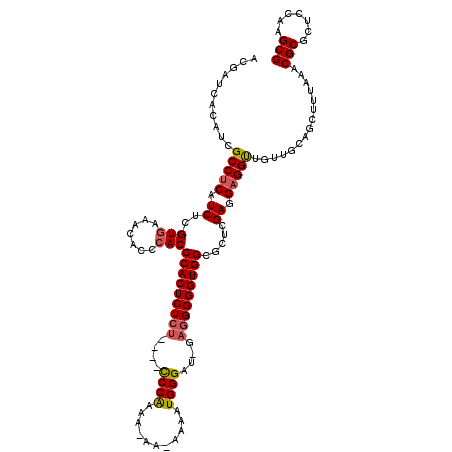
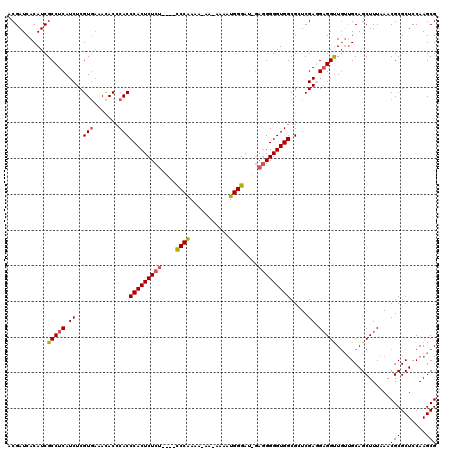
| Location | 14,942,789 – 14,942,904 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -42.34 |
| Consensus MFE | -29.30 |
| Energy contribution | -30.70 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14942789 115 + 23771897 GCCACCCCCUC-AUCCCAUUUAAUUUUUUUGGG----AGAGAGUGGGUGGGUGUUUCACGAGAUGAGGCGAUGUGAUCGUUAACGGUUUACAUUGAGCACUGACUCGGCAAUCGGCGGAU .((((((.(((-.(((((...........))))----)..))).))))))....(((.((((.(.((.(((((((((((....))).))))))))....)).)))))((.....))))). ( -41.60) >DroSec_CAF1 123115 114 + 1 GCCACCCCCUC-AUCCCAUUUU-UUAUUUUGGG----AGAGAGUGGGUGGGUGUUUCACGAGAUGAGGCGAUGUGAUCGUUAACGGUUUACAUUGAGCACUGACUCGACGAUCGACGGAU .((((((.(((-.(((((....-......))))----)..))).)))))).((((((.((((.(.((.(((((((((((....))).))))))))....)).)))))..))..))))... ( -40.30) >DroSim_CAF1 120877 114 + 1 GCCACCCCCUC-AUCCCAUUUU-UUAUUUUGGG----AGAGAGUGGGUGGGUGUUUUACGAGAUGAGGCGAUGUGAUCGUUAACGGUUUACAUUGAGCACUGACUCGACGAUCGGCGGAU .((((((.(((-.(((((....-......))))----)..))).))))))........((((.(.((.(((((((((((....))).))))))))....)).)))))............. ( -40.10) >DroEre_CAF1 117879 107 + 1 GCCACCCC---GCUCCCAAUUC-----UUCGGG----AGAGAGUGGGUG-GUGUUGCACGAGAUGAGGCGAUGUGAUCGUUAACGGUUUACAUUGAGCACUGACUCGCCGAUCGCCGGAU ((((((((---.(((((.....-----...)))----))...).)))))-))......((.(((..(((((((((((((....))).)))))))(((......)))))).)))..))... ( -41.70) >DroYak_CAF1 118997 115 + 1 GCCACCCCCUCUCUCCCAAUUC-----UUUGGAAGAGAGAGAGUGGGUGGGUAUUUCACGAGAUGAGGCGAUGUGAUCGGUAACGGUUUACAUUGAGCACUGACUCUGCCAUCGUCGGAU .((((((((((((((((((...-----.))))..))))))).).))))))...........((((((((((((((((((....))).)))))))(((......))).))).))))).... ( -48.00) >consensus GCCACCCCCUC_AUCCCAUUUC_UU_UUUUGGG____AGAGAGUGGGUGGGUGUUUCACGAGAUGAGGCGAUGUGAUCGUUAACGGUUUACAUUGAGCACUGACUCGACGAUCGGCGGAU .((((((((((...((((...........)))).....))).).))))))........((((.(.((.(((((((((((....))).))))))))....)).)))))............. (-29.30 = -30.70 + 1.40)



| Location | 14,942,789 – 14,942,904 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14942789 115 - 23771897 AUCCGCCGAUUGCCGAGUCAGUGCUCAAUGUAAACCGUUAACGAUCACAUCGCCUCAUCUCGUGAAACACCCACCCACUCUCU----CCCAAAAAAAUUAAAUGGGAU-GAGGGGGUGGC .......(((((..((((....))))((((.....))))..)))))...((((........)))).....((((((.(.((((----((((...........))))).-)))))))))). ( -31.20) >DroSec_CAF1 123115 114 - 1 AUCCGUCGAUCGUCGAGUCAGUGCUCAAUGUAAACCGUUAACGAUCACAUCGCCUCAUCUCGUGAAACACCCACCCACUCUCU----CCCAAAAUAA-AAAAUGGGAU-GAGGGGGUGGC ....((.((((((.((((....))))((((.....)))).)))))))).((((........)))).....((((((.(.((((----((((......-....))))).-)))))))))). ( -35.10) >DroSim_CAF1 120877 114 - 1 AUCCGCCGAUCGUCGAGUCAGUGCUCAAUGUAAACCGUUAACGAUCACAUCGCCUCAUCUCGUAAAACACCCACCCACUCUCU----CCCAAAAUAA-AAAAUGGGAU-GAGGGGGUGGC ..(((((((((((.((((....))))((((.....)))).))))))......(((((((((((....................----..........-...)))))))-)))).))))). ( -32.91) >DroEre_CAF1 117879 107 - 1 AUCCGGCGAUCGGCGAGUCAGUGCUCAAUGUAAACCGUUAACGAUCACAUCGCCUCAUCUCGUGCAACAC-CACCCACUCUCU----CCCGAA-----GAAUUGGGAGC---GGGGUGGC ...(((.(((.(((((....(((.((((((.....))))...)).))).)))))..)))))).......(-(((((.....((----(((((.-----...))))))).---.)))))). ( -34.50) >DroYak_CAF1 118997 115 - 1 AUCCGACGAUGGCAGAGUCAGUGCUCAAUGUAAACCGUUACCGAUCACAUCGCCUCAUCUCGUGAAAUACCCACCCACUCUCUCUCUUCCAAA-----GAAUUGGGAGAGAGGGGGUGGC .(((((.((((((.((((....))))...((((....))))..........)))..)))))).)).........((((((((((((((((((.-----...)))))))))))))))))). ( -43.50) >consensus AUCCGCCGAUCGCCGAGUCAGUGCUCAAUGUAAACCGUUAACGAUCACAUCGCCUCAUCUCGUGAAACACCCACCCACUCUCU____CCCAAAA_AA_AAAAUGGGAU_GAGGGGGUGGC .......(((((..((((....))))((((.....))))..)))))...((((........)))).........(((((((((....((((...........))))....))))))))). (-24.62 = -25.18 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:01 2006