| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,941,385 – 14,941,493 |
| Length | 108 |
| Max. P | 0.832721 |

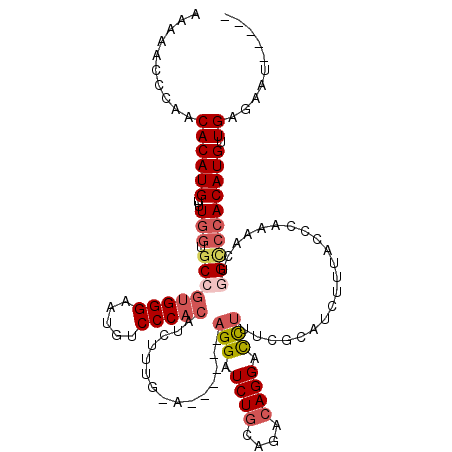
| Location | 14,941,385 – 14,941,493 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14941385 108 + 23771897 -----AUUCUCAACAUGUGGGCCAGUUUUGGGUAAAGAUGCGAAAGGUCCUGUCUGCAGAUCCU------U-CAAAGAUGUGGGACAUUCCCACGGCACCAAACCAUGUGUUGGGUUUUU -----...(((((((((((((((.......)))...(.((((((.((..(((....)))..)))------)-).....((((((.....))))))))).)...))))))))))))..... ( -38.90) >DroSec_CAF1 121695 107 + 1 -----AUUCUCAACAUGUGGGCCAGUUUUGGGUUAGGAUGCGAUAGGUCCUGUCUGCAGAUCCU------U-CAAAGAUGUGGGACAUUCCCACGGCACCACA-CAUGUGUUGGGUUUUU -----...(((((((((((((((..((((((...((((((((((((...))))).)))..))))------)-)))))..(((((.....)))))))).)))))-....)))))))..... ( -41.00) >DroSim_CAF1 119439 107 + 1 -----ACUCUCAACAUGUGGGCUAGUUUUGGGUAAAGAUGCGAAAGAUCCUGUCUGCAGAUCCU------U-CAAAGAUGUGGGACAUUCCCACGGCACCAAA-CAUGUGUUGGGUUUUU -----...(((((((((((.(((.......)))...(.((((((.((((.((....)))))).)------)-).....((((((.....))))))))).)...-)))))))))))..... ( -33.20) >DroEre_CAF1 116442 107 + 1 -----UUUCUCAACAUGUGGGCCUGCUUUGGGUAAGGAUGCGAAAGGUCCUGUCUGCAGAUCCU------U-CAAAGAUGUGGGACAUUCCCACGGCACCAAA-CAUGUGUUGGGUUUUU -----...(((((((((((.((((.....))))..((.((((((.((..(((....)))..)))------)-).....((((((.....)))))))))))...-)))))))))))..... ( -40.10) >DroYak_CAF1 117404 107 + 1 -----AUUCUCAGCAUGUGGUCUUGCUUUGGGUUAAGAUGCGAAAGGUCCUGUCUGCAGAUCCU------U-CAAAGAUGUGGGACAUUCCCACGGCACCAAA-CAUGUGUUGGGUUUUU -----...((((((((((((((((((.....).))))))(((((.((..(((....)))..)))------)-).....((((((.....))))))))......-)))))))))))..... ( -35.80) >DroAna_CAF1 106693 113 + 1 AUUUUUUUCACAGCAUGUCUACCACAU---GGUCCUAA--CUCACAAUCCUUACUUCAGAUCCUGUGGUAUCUGAGAAUGUGGGACAUUCCCUUGGCAG-AAA-CAUGUGUUGGGCUUAU ................(((((.(((((---(((((...--..((((.((......((((((((...)).)))))))).)))))))).(((........)-)).-)))))).))))).... ( -27.40) >consensus _____AUUCUCAACAUGUGGGCCAGUUUUGGGUAAAGAUGCGAAAGGUCCUGUCUGCAGAUCCU______U_CAAAGAUGUGGGACAUUCCCACGGCACCAAA_CAUGUGUUGGGUUUUU ........(((((((((((.(((.......))).....(((....((((.((....))))))................((((((.....)))))))))......)))))))))))..... (-23.88 = -24.30 + 0.42)

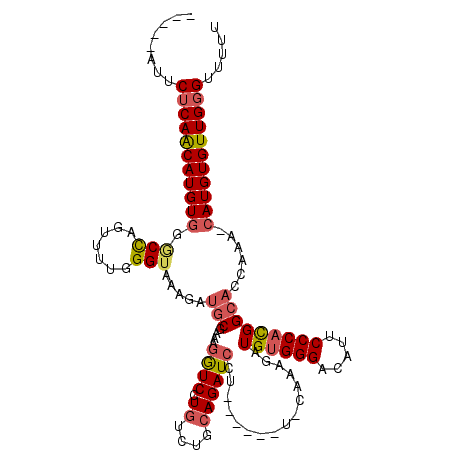

| Location | 14,941,385 – 14,941,493 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -18.37 |
| Energy contribution | -19.40 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14941385 108 - 23771897 AAAAACCCAACACAUGGUUUGGUGCCGUGGGAAUGUCCCACAUCUUUG-A------AGGAUCUGCAGACAGGACCUUUCGCAUCUUUACCCAAAACUGGCCCACAUGUUGAGAAU----- .......(((((..(((...(((((.(((((.....)))))......(-(------(((.((((....)))).))))).))))).....(((....))).)))..))))).....----- ( -32.10) >DroSec_CAF1 121695 107 - 1 AAAAACCCAACACAUG-UGUGGUGCCGUGGGAAUGUCCCACAUCUUUG-A------AGGAUCUGCAGACAGGACCUAUCGCAUCCUAACCCAAAACUGGCCCACAUGUUGAGAAU----- .......(((((....-(((((.((((((((.....)))))......(-(------(((.((((....)))).))).))..................))))))))))))).....----- ( -33.20) >DroSim_CAF1 119439 107 - 1 AAAAACCCAACACAUG-UUUGGUGCCGUGGGAAUGUCCCACAUCUUUG-A------AGGAUCUGCAGACAGGAUCUUUCGCAUCUUUACCCAAAACUAGCCCACAUGUUGAGAGU----- ...........(((((-(..((.((.(((((.....)))))......(-(------(((((((.......)))))))))...................)))))))))).......----- ( -28.90) >DroEre_CAF1 116442 107 - 1 AAAAACCCAACACAUG-UUUGGUGCCGUGGGAAUGUCCCACAUCUUUG-A------AGGAUCUGCAGACAGGACCUUUCGCAUCCUUACCCAAAGCAGGCCCACAUGUUGAGAAA----- ...........(((((-(..((.((((((((.....)))))..(((((-(------((((..(((.((.((...)).))))))))))...)))))..))))))))))).......----- ( -31.40) >DroYak_CAF1 117404 107 - 1 AAAAACCCAACACAUG-UUUGGUGCCGUGGGAAUGUCCCACAUCUUUG-A------AGGAUCUGCAGACAGGACCUUUCGCAUCUUAACCCAAAGCAAGACCACAUGCUGAGAAU----- ...............(-((.(((((.(((((.....)))))......(-(------(((.((((....)))).))))).)))))..)))....((((........))))......----- ( -26.50) >DroAna_CAF1 106693 113 - 1 AUAAGCCCAACACAUG-UUU-CUGCCAAGGGAAUGUCCCACAUUCUCAGAUACCACAGGAUCUGAAGUAAGGAUUGUGAG--UUAGGACC---AUGUGGUAGACAUGCUGUGAAAAAAAU .........(((((((-((.-.......(((((((.....)))))))...(((((((((.(((((..((.......))..--))))).))---.))))))))))))).)))......... ( -30.90) >consensus AAAAACCCAACACAUG_UUUGGUGCCGUGGGAAUGUCCCACAUCUUUG_A______AGGAUCUGCAGACAGGACCUUUCGCAUCUUUACCCAAAACUGGCCCACAUGUUGAGAAU_____ ..........((((((...(((.((((((((.....)))))...............(((.((((....)))).))).....................)))))))))).)).......... (-18.37 = -19.40 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:55 2006