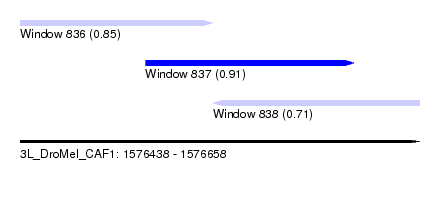
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,576,438 – 1,576,658 |
| Length | 220 |
| Max. P | 0.906617 |

| Location | 1,576,438 – 1,576,544 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1576438 106 + 23771897 AACGAAUAAUAGCGGUGCACGUAAAAAGGGAAAACGAAUAAAUCUACGAACAACGAACUGCAAGCGAGAGCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUUCGUC .(((((.....(((((...(((......(((...........))).......))).)))))..((....))((((((((....)))))))).........))))). ( -25.02) >DroSec_CAF1 20663 106 + 1 AACGAAUAAUAGCGGUGCACGUGAAAAGGGAAAACGAAUAAAUCUACGAACAACGAACUGCAAGCGAGAGCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUUCGUC .(((((.....(((((...(((......(((...........))).......))).)))))..((....))((((((((....)))))))).........))))). ( -26.12) >DroSim_CAF1 21039 106 + 1 AACGAAUAAUAGCGGUGCACUUGGAAAGGGAAAACGAAUAAAUCUACGAAUAACGAACUGCAAGCGAGAGCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUUCGUC .(((((.....(((((...(((....))).................((.....)).)))))..((....))((((((((....)))))))).........))))). ( -25.30) >DroEre_CAF1 27775 106 + 1 AACGAAUAAUAGCGGUGCAUGCAAAAAGGGAAAACGAAUAAAUCUACGAACAACGAACUGCAAGCGAGAGCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUGCGUU ...........(((((((.((((....(......)...........((.....))...)))).((....))((((((((....))))))))....))).))))... ( -23.60) >DroYak_CAF1 21790 106 + 1 AACGAAUAAUAGCGGUGCAUGCAAAAAGGGAAAACGAAUAAAUCUACGAACAACGAACUGCAAGCGAGAGCGAGAUAGCAACCGCUGUCUUUUUAGCAUUUGCGAC ...(((..((((((((((.((((....(......)...........((.....))...)))).((....))......)).))))))))..)))..((....))... ( -23.50) >consensus AACGAAUAAUAGCGGUGCACGUAAAAAGGGAAAACGAAUAAAUCUACGAACAACGAACUGCAAGCGAGAGCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUUCGUC .(((((.....(((((............(((...........)))...........)))))..((....))((((((((....)))))))).........))))). (-20.64 = -21.24 + 0.60)

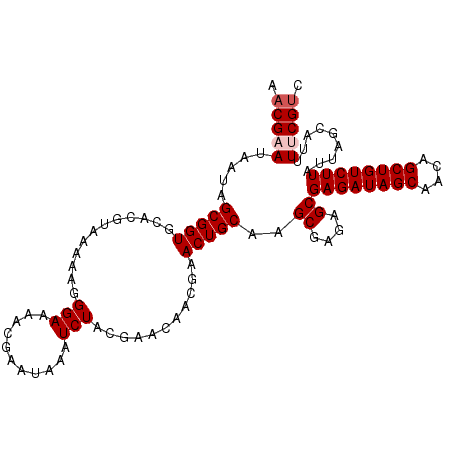
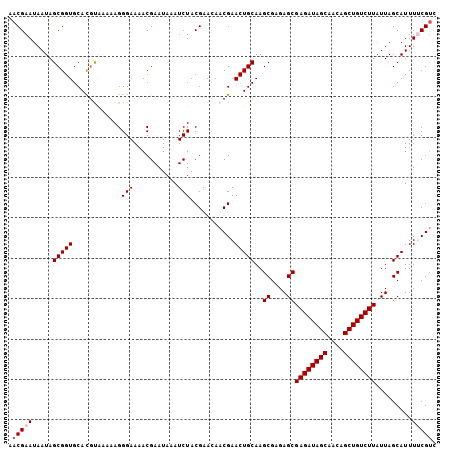
| Location | 1,576,507 – 1,576,622 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.88 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -12.95 |
| Energy contribution | -13.76 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1576507 115 + 23771897 GCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUU-CGU--CGCGGGGGAGUAGAAGUAAAAGUGCACUC-AAAGAACGGAGAGAGU-GCAGUGUGAAAGAGAGAGCAAGCAGUUGGUCUUG ..((((((((....))))))))....(((((((-(..--((.....((((....(((....)))))))-......))..))))))-))...........((((.(((....))).)))). ( -31.20) >DroSec_CAF1 20732 113 + 1 GCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUU-CGU--CGCGGGGGAGUAGAAGUAAAAGUGCACUC-CAAGAACGGAGAGAGU-GCAGU--GAAAGAGAGAGCAAGCAGUUAGUCUUG ..((((((((....))))))))....(((((((-(..--((....(((((....(((....)))))))-).....))..))))))-))...--....((((.(((.....)))..)))). ( -32.80) >DroSim_CAF1 21108 113 + 1 GCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUU-CGU--CGCGGGGGAGUAGAAGUAAGAAUGCACUC-CAAGAACGGAGAGAGU-GCAGU--GAAAGAGAGAGCAAGCAGUUAGUCUUG ..((((((((....))))))))....(((((((-(..--((....(((((....(((....)))))))-).....))..))))))-))...--....((((.(((.....)))..)))). ( -32.80) >DroEre_CAF1 27844 106 + 1 GCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUG-CGU--UGCGGGGGAGUAGAAGUAAAAGUGCACUC-AAAGAUAAGAGAUUGU-GCAGU--GAAAGAGAAAGCAAGCAGUU------- ((((((((((....))))))))....((.((((-(.(--(((......))))..))))).(..(((((-........)))..)).-.)...--..........))..))....------- ( -25.40) >DroYak_CAF1 21859 87 + 1 GCGAGAUAGCAACCGCUGUCUUUUUAGCAUUUG-CGA--CGCGGGGGAGUAGAAGUAAAAGUGCACUC-AAUGAUAAGAGAGUGU-GC---------------------AGUU------- ..((((((((....))))))))(((.((.((((-(..--(.....)..))))).)).)))(..(((((-..........))))).-.)---------------------....------- ( -25.20) >DroAna_CAF1 29405 101 + 1 GUAAGAUAGUCG-----CUCUUAUUAGCAUUGCAAGACACUCGGGAGA------GUAAAAGUGCAGCAGGUUGACCAGAGAAAGAAGGAGA--GAGAGUGAUAAGAAGCAGUUA------ ........((((-----(((((.(((((.(((((....((((....))------)).....)))))...)))))((..........))...--)))))))))............------ ( -25.10) >consensus GCGAGAUAGCAACAGCUGUCUUAUUAGCAUUUU_CGU__CGCGGGGGAGUAGAAGUAAAAGUGCACUC_AAAGAACAGAGAGAGU_GCAGU__GAAAGAGAGAGCAAGCAGUUA______ ((((((((((....))))))))....((((((........((......))........))))))......................))................................ (-12.95 = -13.76 + 0.81)

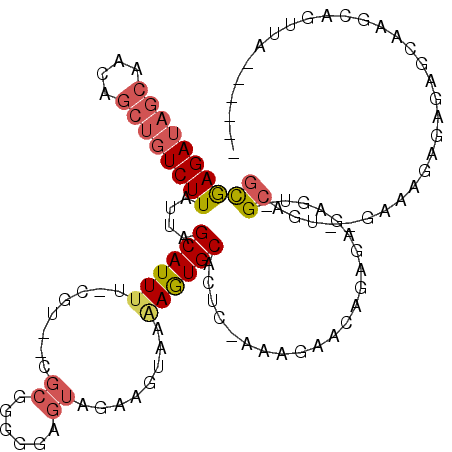
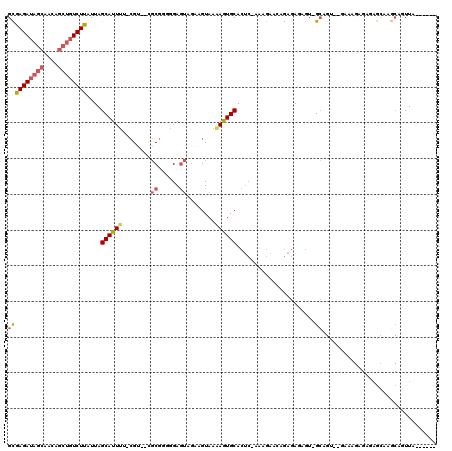
| Location | 1,576,544 – 1,576,658 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -17.29 |
| Consensus MFE | -9.48 |
| Energy contribution | -10.67 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
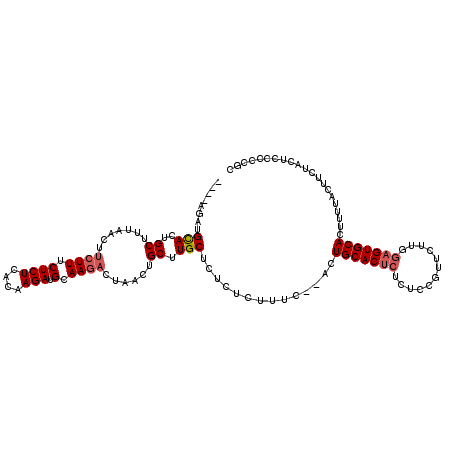
>3L_DroMel_CAF1 1576544 114 - 23771897 ----AGAUGCACUGCUUUAACUUCUUUCUCUCACAAGAUGCAAGACCAACUGCUUGCUCUCUCUUUCACACUGCACUCUCUCCGUUCUUUGAGUGCACUUUUACUUCUACUCCCCCGC ----....((.........................(((.(((((........))))))))...........(((((((............)))))))...................)) ( -17.40) >DroSec_CAF1 20769 112 - 1 ----AUAUGUACUGCUUUAACUUCUUUCUCUCACAAGAUGCAAGACUAACUGCUUGCUCUCUCUUUC--ACUGCACUCUCUCCGUUCUUGGAGUGCACUUUUACUUCUACUCCCCCGC ----....(((........................(((.(((((.(.....))))))))).......--..((((((((..........))))))))....))).............. ( -17.50) >DroSim_CAF1 21145 112 - 1 ----AGAUGCACUGCUUUAACUUCUUGCUCUCACAAGAUGCAAGACUAACUGCUUGCUCUCUCUUUC--ACUGCACUCUCUCCGUUCUUGGAGUGCAUUCUUACUUCUACUCCCCCGC ----(((.(((..((.......(((((((((....))).))))))......)).)))))).......--..((((((((..........))))))))..................... ( -24.32) >DroEre_CAF1 27881 93 - 1 UCGAACUGGCACUGCUUUAACUCC-----------------------AACUGCUUGCUUUCUCUUUC--ACUGCACAAUCUCUUAUCUUUGAGUGCACUUUUACUUCUACUCCCCCGC ..((...((((..((.........-----------------------....)).))))...))....--..(((((....((........)))))))..................... ( -9.92) >consensus ____AGAUGCACUGCUUUAACUUCUUUCUCUCACAAGAUGCAAGACUAACUGCUUGCUCUCUCUUUC__ACUGCACUCUCUCCGUUCUUGGAGUGCACUUUUACUUCUACUCCCCCGC ........(((..((.......((((.((((....))).).))))......)).)))..............(((((((............)))))))..................... ( -9.48 = -10.67 + 1.19)

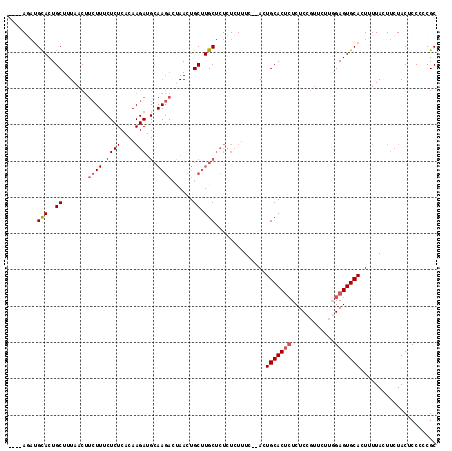
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:02 2006