
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,936,814 – 14,936,919 |
| Length | 105 |
| Max. P | 0.933077 |

| Location | 14,936,814 – 14,936,919 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.01 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -30.65 |
| Energy contribution | -30.46 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14936814 105 - 23771897 --------UAG---AUGGAGUG--GGA-UAGGAUCCUGGCCCAUCCACUUGGGGGGUGACCAACCAGGAACCUAGCGUGCUUGUGGCUCGCUUAACUAUUCCACAGGCAUAAUUCCCCA --------...---.....(.(--(((-((((.((((((((((......))))((....))..)))))).))))..((((((((((..............))))))))))...))))). ( -45.94) >DroSec_CAF1 117162 105 - 1 --------UAG---AUGGAGUG--GGA-UUGGAUCCUGGCCCAUCCACUUGGGGGGUGACCAACCAGGAACCUCGCGUGUUUGUGGCUCGCUUAACUAUUCCACAGGCAUAAUUCCCUA --------...---....((.(--(((-((((.((((((((((......))))((....))..)))))).))....((((((((((..............)))))))))))))))))). ( -40.64) >DroSim_CAF1 114683 105 - 1 --------UAG---AUGGAGUG--GGA-UAGGAUCCUGGCCCAUCCACUUGGGGGGUGACCAACCAGGAACCUCGCGUGUUUGUGGCUCGCUUAACUAUUCCACAGGCAUAAUUCCCUA --------...---.......(--(((-.(((.((((((((((......))))((....))..)))))).)))...((((((((((..............))))))))))...)))).. ( -41.14) >DroEre_CAF1 111918 113 - 1 CUUCGUUGUGU---AUGGAGUG--GGA-UAGGAUCCUGGCCCAUCCACUUGGGGGGCGCACAACCAGGAACCUCACAUGUUUGUGGCUCGCUUAACUAUUCCACAGGCAUAAUUCCUUA ....(((((((---.(((((((--(..-.(((.(((((((((.(((....))))))).......))))).)))((((....))))..........))))))))...)))))))...... ( -38.31) >DroYak_CAF1 112725 113 - 1 CGUCGUUGUGG---AUGGAGUG--AGA-UAGUAUCCUGGCCCAUCCACUUGGGGGGUGCUCAGCCAGGAACCUCGCAUGUUUGUGGCUCGCUUAACUAUUCCACAGGCAUAAUUCCCUA .......((((---((((.((.--((.-.......)).)))))))))).((((((((.....((.((....)).))((((((((((..............)))))))))).)))))))) ( -41.04) >DroPer_CAF1 165969 119 - 1 UGACGUUGGGUAGGGUAGAGUACCAGAUCAAGAUCCUUUUCCAUCUACUUGGGGGGUGGACAGCGAAGGGCUUCGAAUGGUUUUGGCUUGUUUAACAAUUCCAUAAGAAUAAUUCUGUA ....((((..((((.((((..((((..(((((.(((((((((((((.......)))))))....))))))))).)).)))))))).))))..)))).((((.....))))......... ( -30.80) >consensus ________UAG___AUGGAGUG__GGA_UAGGAUCCUGGCCCAUCCACUUGGGGGGUGACCAACCAGGAACCUCGCAUGUUUGUGGCUCGCUUAACUAUUCCACAGGCAUAAUUCCCUA ................(((((........(((.((((((((((......))))((....))..)))))).)))...((((((((((..............)))))))))).)))))... (-30.65 = -30.46 + -0.19)

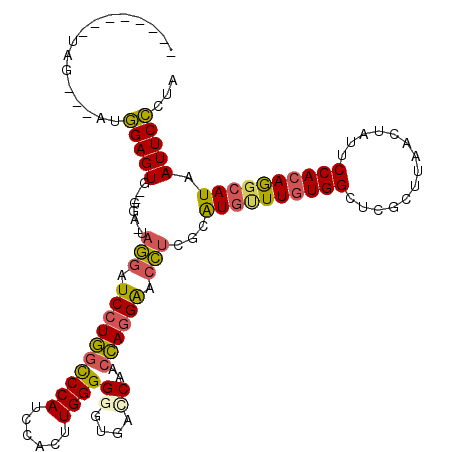

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:48 2006