| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,927,746 – 14,927,843 |
| Length | 97 |
| Max. P | 0.897342 |

| Location | 14,927,746 – 14,927,843 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -19.43 |
| Energy contribution | -21.93 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
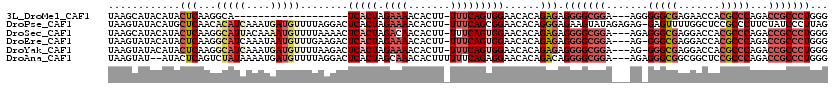
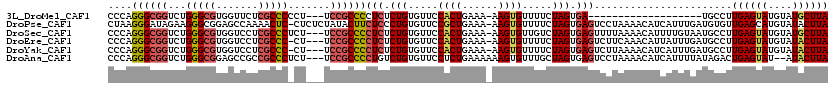
>3L_DroMel_CAF1 14927746 97 + 23771897 UAAGCAUACAUACUCAAGGCA-------------------UCACUAGAAAACACUU-UUUCAGUGGAACACAGAGAGGGGCGGA---AGGGGGCGAGAACCACGCCCAGACCGCCCUGGG ............(((...(..-------------------(((((.(((((....)-)))))))))..)...))).(((((((.---...(((((.......)))))...)))))))... ( -33.50) >DroPse_CAF1 165609 118 + 1 UAAGUAUACAUGCUCAACACAUCAAAUGAUGUUUUAGGACUCACUAGAAAACACUU-UUUCAGCGGAACACAGGGAGAAGUAUAGAGAG-GAGUUUUGGCUCCGCCCUUCUAUCCCUUAG ...((.....((((....(((((....)))))(((((......)))))........-....))))....))(((((..((...((.(.(-((((....))))).).)).)).)))))... ( -25.60) >DroSec_CAF1 108196 116 + 1 UAAGCAUACAUACUCAAGGCAUUACAAAAUGUUUUAAAACUCACUAGACAACACUU-UUUCAGUGGAACACAGAGAGGGGCGGA---AGAGGGCGAGGACCACGCCCAGACCGCCCUGGG ............(((((((((((....)))))))).....(((((.((.((....)-).)))))))......))).(((((((.---...(((((.......)))))...)))))))... ( -33.40) >DroEre_CAF1 103453 115 + 1 UAAGUAUACAUACUCAAGGCAUCAAAUAAUGUUUGAAGACUCACUAGAAAACACUU-UUUCAGUGGAACACAGAGAGGGGCGGA---AG-GGGCGAGGACCACGCCCAGACCGCCCUGGG ............(((.((((((......))))))......(((((.(((((....)-)))))))))......))).(((((((.---..-(((((.......)))))...)))))))... ( -36.80) >DroYak_CAF1 103639 115 + 1 UAAGUAUACAUACUCAAGGCAUCAAAUGAUGUUUUAAGACUCACUAGAAAACACUU-UUUCAGUGGAACACAGAGAGGGGCGGA---AG-GGGCGAGGACCACGCCCAGACCGCCCUGGG ............(((((((((((....)))))))).....(((((.(((((....)-)))))))))......))).(((((((.---..-(((((.......)))))...)))))))... ( -41.50) >DroAna_CAF1 94645 115 + 1 UAAGUAU--AUACUCAGUCUAUAAAAUGAUGUUUUAGGACUCACUAGCAAACACUUUUUUCAGAGGAACACAGACAGGGGCGGA---AGAGGGCGGCGGCUCCGCCCAGACCGCCCUGGG .......--...(((.((((.........((((((((......)))..)))))((((.....)))).....)))).(((((((.---...((((((.....))))))...)))))))))) ( -35.50) >consensus UAAGUAUACAUACUCAAGGCAUCAAAUGAUGUUUUAAGACUCACUAGAAAACACUU_UUUCAGUGGAACACAGAGAGGGGCGGA___AG_GGGCGAGGACCACGCCCAGACCGCCCUGGG ............(((...(((((....)))))........(((((.((((.......)))))))))......))).(((((((.......(((((.......)))))...)))))))... (-19.43 = -21.93 + 2.50)
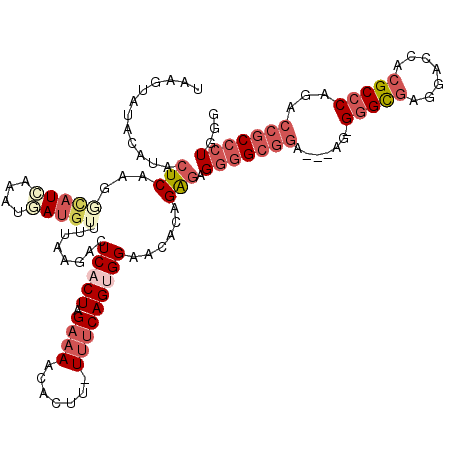
| Location | 14,927,746 – 14,927,843 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
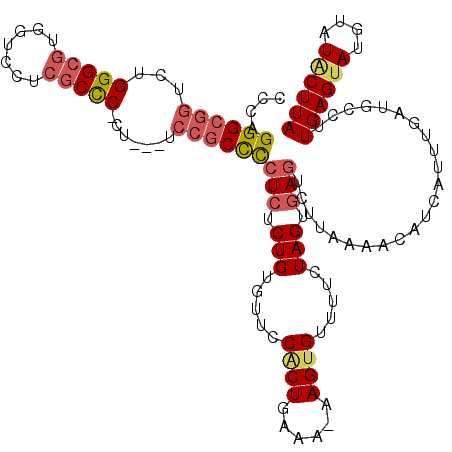
>3L_DroMel_CAF1 14927746 97 - 23771897 CCCAGGGCGGUCUGGGCGUGGUUCUCGCCCCCU---UCCGCCCCUCUCUGUGUUCCACUGAAA-AAGUGUUUUCUAGUGA-------------------UGCCUUGAGUAUGUAUGCUUA ....((((((...(((((.......)))))...---.))))))(((...(.((..((((((((-(....))))).)))).-------------------.)))..)))............ ( -32.70) >DroPse_CAF1 165609 118 - 1 CUAAGGGAUAGAAGGGCGGAGCCAAAACUC-CUCUCUAUACUUCUCCCUGUGUUCCGCUGAAA-AAGUGUUUUCUAGUGAGUCCUAAAACAUCAUUUGAUGUGUUGAGCAUGUAUACUUA ...(((((.((.((((.((((......)))-).))))...))..)))))((((((((((((((-(....))))).)))).........(((((....)))))...))))))......... ( -31.90) >DroSec_CAF1 108196 116 - 1 CCCAGGGCGGUCUGGGCGUGGUCCUCGCCCUCU---UCCGCCCCUCUCUGUGUUCCACUGAAA-AAGUGUUGUCUAGUGAGUUUUAAAACAUUUUGUAAUGCCUUGAGUAUGUAUGCUUA ....((((((...(((((.......)))))...---.))))))(((.(((..(..((((....-.))))..)..))).)))........((((....))))...((((((....)))))) ( -35.90) >DroEre_CAF1 103453 115 - 1 CCCAGGGCGGUCUGGGCGUGGUCCUCGCCC-CU---UCCGCCCCUCUCUGUGUUCCACUGAAA-AAGUGUUUUCUAGUGAGUCUUCAAACAUUAUUUGAUGCCUUGAGUAUGUAUACUUA ....((((((...(((((.......)))))-..---.))))))(((.(((.....((((....-.)))).....))).)))...(((((.....))))).....((((((....)))))) ( -34.60) >DroYak_CAF1 103639 115 - 1 CCCAGGGCGGUCUGGGCGUGGUCCUCGCCC-CU---UCCGCCCCUCUCUGUGUUCCACUGAAA-AAGUGUUUUCUAGUGAGUCUUAAAACAUCAUUUGAUGCCUUGAGUAUGUAUACUUA ....((((((...(((((.......)))))-..---.))))))(((.(((.....((((....-.)))).....))).))).(((((..((((....))))..)))))............ ( -37.30) >DroAna_CAF1 94645 115 - 1 CCCAGGGCGGUCUGGGCGGAGCCGCCGCCCUCU---UCCGCCCCUGUCUGUGUUCCUCUGAAAAAAGUGUUUGCUAGUGAGUCCUAAAACAUCAUUUUAUAGACUGAGUAU--AUACUUA ..((((((((...((((((.....))))))...---.))).)))))..................((((((.((((....((((.(((((.....)))))..)))).)))).--)))))). ( -37.60) >consensus CCCAGGGCGGUCUGGGCGUGGUCCUCGCCC_CU___UCCGCCCCUCUCUGUGUUCCACUGAAA_AAGUGUUUUCUAGUGAGUCUUAAAACAUCAUUUGAUGCCUUGAGUAUGUAUACUUA ....((((((...(((((.......))))).......))))))(((.(((.....((((......)))).....))).))).......................((((((....)))))) (-23.74 = -24.55 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:41 2006