| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,575,554 – 1,575,651 |
| Length | 97 |
| Max. P | 0.727888 |

| Location | 1,575,554 – 1,575,651 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
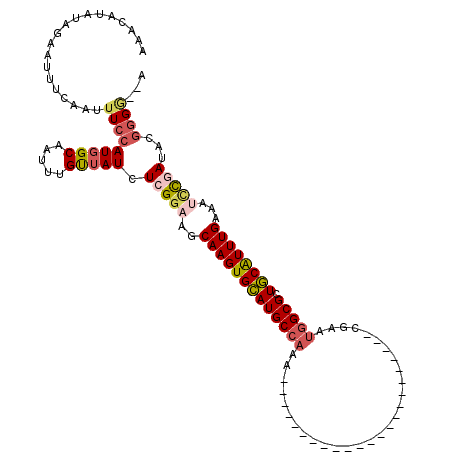
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -15.75 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1575554 97 + 23771897 UCUGCCCGUAUCAGAUUUCAAAUGCAGCGCCAUUCG-----------------------UUUGGCAUGCACUUGCUUCCGAGAUAACAAAUUGCCAUGGAAAUUGAAAUUCUAUAUGUUU ......(((((..((((((((.((((..((((....-----------------------..)))).)))).....(((((.(.(((....))).).))))).))))))))..)))))... ( -23.30) >DroSec_CAF1 19818 97 + 1 UCUUCCCGUAUCAGAUUUCAAAUGCAGCGCCAUUCG-----------------------UUUGGCAUGCACUUGCUUCCGAGAUAACAAAUUGCCAUGGAAAUUGAAAUUCUAUAUGUUU ......(((((..((((((((.((((..((((....-----------------------..)))).)))).....(((((.(.(((....))).).))))).))))))))..)))))... ( -23.30) >DroSim_CAF1 20182 97 + 1 UCUUCCCGUAUCAGAUUUCAAAUGCAGCGCCAUUCG-----------------------UUUGGCAUGCACUUGCUUCCGAGAUAACAAAUUGCCAUGGAAAUUGAAAUUCUAUAUGUUU ......(((((..((((((((.((((..((((....-----------------------..)))).)))).....(((((.(.(((....))).).))))).))))))))..)))))... ( -23.30) >DroEre_CAF1 26854 95 + 1 U--CCCCAUAUCGUAUUUCAAACGCAGCGCCACUCG-----------------------CUUGGCAUGCACUUGCUUCCGAGAUAACAAAUUGCCAUGGAAAUGGAAAUUCCAUAUGCUU .--...(((((.(.(((((....(((..((((....-----------------------..)))).))).((((....))))............(((....)))))))).).)))))... ( -19.40) >DroYak_CAF1 20893 95 + 1 U--CCCCAUAUUGAAUUUCAAACGCAGCGCCACUCG-----------------------CCUGGCAUGCACUUGCUACCGAGAUAGCAAAUUGCCAUGGAAACGGAAAUUCUAUAUGUUU .--...(((((.(((((((....(((..((((....-----------------------..)))).)))..((((((......)))))).......((....))))))))).)))))... ( -27.10) >DroAna_CAF1 28462 118 + 1 U--CCUCGUAUUGAGUUUCAAACGAAGCGCCCUGCCUGCCGGCCAGUUGCCAGUUGCCAGUUUGCAUUCACUUGCUUCCGAGAUAACAAAUUGCAAUUGAAAUUGAAAUUCUAUAUGUUU .--...(((((.(((((((((.....(((.(..(((....)))..).)))(((((((.((((((......((((....))))....)))))))))))))...))))))))).)))))... ( -30.90) >consensus U__CCCCGUAUCAGAUUUCAAACGCAGCGCCAUUCG_______________________UUUGGCAUGCACUUGCUUCCGAGAUAACAAAUUGCCAUGGAAAUUGAAAUUCUAUAUGUUU ......(((((..((((((((..(((..((((.............................)))).))).((((....))))....................))))))))..)))))... (-15.75 = -15.95 + 0.20)

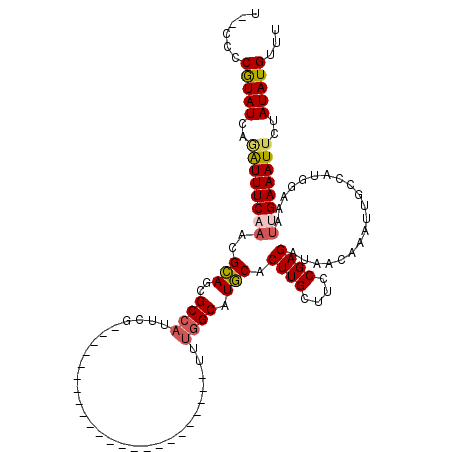
| Location | 1,575,554 – 1,575,651 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1575554 97 - 23771897 AAACAUAUAGAAUUUCAAUUUCCAUGGCAAUUUGUUAUCUCGGAAGCAAGUGCAUGCCAAA-----------------------CGAAUGGCGCUGCAUUUGAAAUCUGAUACGGGCAGA .....(((.(((.......))).)))....((((((...(((((..(((((((((((((..-----------------------....))))).))))))))...)))))....)))))) ( -25.60) >DroSec_CAF1 19818 97 - 1 AAACAUAUAGAAUUUCAAUUUCCAUGGCAAUUUGUUAUCUCGGAAGCAAGUGCAUGCCAAA-----------------------CGAAUGGCGCUGCAUUUGAAAUCUGAUACGGGAAGA ..................((((((((((.....))))).(((((..(((((((((((((..-----------------------....))))).))))))))...)))))...))))).. ( -28.10) >DroSim_CAF1 20182 97 - 1 AAACAUAUAGAAUUUCAAUUUCCAUGGCAAUUUGUUAUCUCGGAAGCAAGUGCAUGCCAAA-----------------------CGAAUGGCGCUGCAUUUGAAAUCUGAUACGGGAAGA ..................((((((((((.....))))).(((((..(((((((((((((..-----------------------....))))).))))))))...)))))...))))).. ( -28.10) >DroEre_CAF1 26854 95 - 1 AAGCAUAUGGAAUUUCCAUUUCCAUGGCAAUUUGUUAUCUCGGAAGCAAGUGCAUGCCAAG-----------------------CGAGUGGCGCUGCGUUUGAAAUACGAUAUGGGG--A ..((.(((((((.......)))))))))...((.((((.(((....(((((((((((((..-----------------------....))))).)))))))).....))).)))).)--) ( -27.90) >DroYak_CAF1 20893 95 - 1 AAACAUAUAGAAUUUCCGUUUCCAUGGCAAUUUGCUAUCUCGGUAGCAAGUGCAUGCCAGG-----------------------CGAGUGGCGCUGCGUUUGAAAUUCAAUAUGGGG--A ...(((((.((((((((((.....((((((((((((((....)))))))))...)))))((-----------------------((.....)))))))...))))))).)))))...--. ( -32.50) >DroAna_CAF1 28462 118 - 1 AAACAUAUAGAAUUUCAAUUUCAAUUGCAAUUUGUUAUCUCGGAAGCAAGUGAAUGCAAACUGGCAACUGGCAACUGGCCGGCAGGCAGGGCGCUUCGUUUGAAACUCAAUACGAGG--A ...................((((.((((..((((......)))).)))).)))).((...(((.(..(((((.....)))))..).))).)).(((((((((.....))).))))))--. ( -29.90) >consensus AAACAUAUAGAAUUUCAAUUUCCAUGGCAAUUUGUUAUCUCGGAAGCAAGUGCAUGCCAAA_______________________CGAAUGGCGCUGCAUUUGAAAUCCGAUACGGGG__A ...................(((((((((.....))))).(((((..(((((((((((((.............................))))).))))))))...)))))...))))... (-15.98 = -16.57 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:59 2006