| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
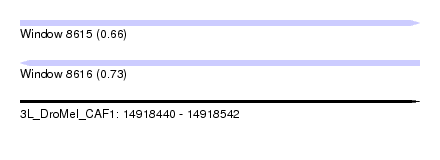
| Location | 14,918,440 – 14,918,542 |
| Length | 102 |
| Max. P | 0.728594 |

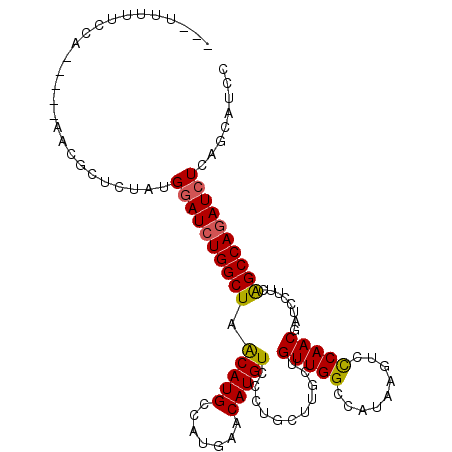
| Location | 14,918,440 – 14,918,542 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -24.37 |
| Energy contribution | -25.23 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14918440 102 + 23771897 GGUUGCUGAGAUCUGGCUGAAGGAUCGUUGGGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAUAGCAUGUUAGCCAGAUCCAUAGAGCGUU-----UGGAAAAA--- .(((.(((.(((((((((........(((((..(....).)))))..........(((((((.....))))))))))))))))..))))))...-----........--- ( -31.30) >DroSec_CAF1 100683 102 + 1 GGAUGCUGAGAUCUGGCUGAAGGAUCGUUGGGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAUAGCAUGUUAGCCAUAUCCACAGAGCGUU-----CGGAAAAA--- (((((((..(((((.......))))).((((((..((((((.((((((.((((......))))....)).)))).))))))))).)))))))))-----).......--- ( -32.10) >DroSim_CAF1 97901 102 + 1 AGUUGCUGAGAUCUGGCUGAAGGAUCGUUGGGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAUAGCAUGUUAGCCAGAUCCACAGAGCGUU-----CGGAAAAA--- .(((.(((.(((((((((........(((((..(....).)))))..........(((((((.....))))))))))))))))..))))))...-----........--- ( -33.40) >DroEre_CAF1 95965 105 + 1 GGAUGCUGAGAUCUGGCUGAAGGAUCGUUGGGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAAGGCAUGUUAGCCAGAUCCAUAGGCCGCU-----UGAAAAAAAUA ((....((.(((((((((........(((((..(....).)))))..........(((((((.....))))))))))))))))))....))...-----........... ( -32.40) >DroYak_CAF1 95505 102 + 1 GGAUGCUGAGAUCUGGCUAAAGGAUCGUUGGGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAAGGCAUGCUAGCCAGAUCCAUAGACAGCU-----UGGAAAAA--- ....((((.((((((((((.......(((((..(....).))))).(((.((..(((....)))...)).)))))))))))))......)))).-----........--- ( -34.00) >DroAna_CAF1 87819 99 + 1 -----CAGAUAUCUGGCCAAGAUCCAGUUGAGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAGGACAUGUUAGCCAGAUUCAGAGAUCCCUGAUCCCGGGG------ -----..((.(((((((......((.((((.(.(....).))))).(....).))((((((((...)))))))).)))))))))......(((((....)))))------ ( -28.30) >consensus GGAUGCUGAGAUCUGGCUGAAGGAUCGUUGGGACUUAUGGCCAACAGCAAGCAGGGACAUGUUCAUAGCAUGUUAGCCAGAUCCAUAGAGCGCU_____CGGAAAAA___ .....(((.(((((((((........(((((..(....).)))))..........(((((((.....))))))))))))))))..)))...................... (-24.37 = -25.23 + 0.86)

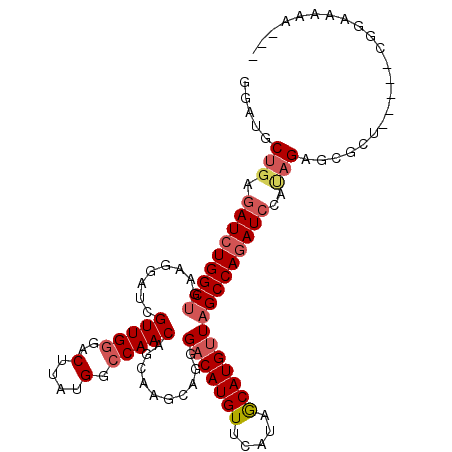
| Location | 14,918,440 – 14,918,542 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -22.07 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14918440 102 - 23771897 ---UUUUUCCA-----AACGCUCUAUGGAUCUGGCUAACAUGCUAUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCCCAACGAUCCUUCAGCCAGAUCUCAGCAACC ---........-----...(((....((((((((((.(((((.......)))))..........((((((.........)))))).......)))))))))).))).... ( -26.90) >DroSec_CAF1 100683 102 - 1 ---UUUUUCCG-----AACGCUCUGUGGAUAUGGCUAACAUGCUAUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCCCAACGAUCCUUCAGCCAGAUCUCAGCAUCC ---........-----...(((.((.((((((((((((((.((...................))))))))))))..))))))..((((.........))))..))).... ( -24.81) >DroSim_CAF1 97901 102 - 1 ---UUUUUCCG-----AACGCUCUGUGGAUCUGGCUAACAUGCUAUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCCCAACGAUCCUUCAGCCAGAUCUCAGCAACU ---........-----...(((....((((((((((.(((((.......)))))..........((((((.........)))))).......)))))))))).))).... ( -26.90) >DroEre_CAF1 95965 105 - 1 UAUUUUUUUCA-----AGCGGCCUAUGGAUCUGGCUAACAUGCCUUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCCCAACGAUCCUUCAGCCAGAUCUCAGCAUCC ...........-----.(((((....((((((((((((((.((...................)))))))))))...((....)))))))....))).(....).)).... ( -25.51) >DroYak_CAF1 95505 102 - 1 ---UUUUUCCA-----AGCUGUCUAUGGAUCUGGCUAGCAUGCCUUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCCCAACGAUCCUUUAGCCAGAUCUCAGCAUCC ---........-----.((((.....((((((((((((((.((...((.....))...)).)))((((((.........))))))......))))))))))))))).... ( -32.10) >DroAna_CAF1 87819 99 - 1 ------CCCCGGGAUCAGGGAUCUCUGAAUCUGGCUAACAUGUCCUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCUCAACUGGAUCUUGGCCAGAUAUCUG----- ------..(((((.(((((....))))).)))))...((((((.....))))))............(((((((...((((.....))))..))))))).......----- ( -27.70) >consensus ___UUUUUCCA_____AACGCUCUAUGGAUCUGGCUAACAUGCCAUGAACAUGUCCCUGCUUGCUGUUGGCCAUAAGUCCCAACGAUCCUUCAGCCAGAUCUCAGCAUCC ..........................((((((((((.(((((.......)))))...........(((((.........)))))........))))))))))........ (-22.07 = -21.98 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:32 2006