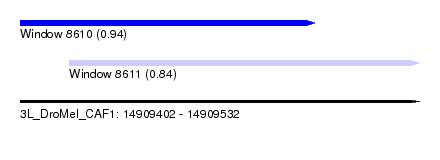
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,909,402 – 14,909,532 |
| Length | 130 |
| Max. P | 0.935709 |

| Location | 14,909,402 – 14,909,498 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -30.26 |
| Energy contribution | -29.98 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
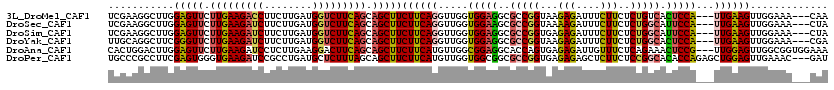
>3L_DroMel_CAF1 14909402 96 + 23771897 GGGAUCGGAAUCUCCUUCGAAGGCUUGGAGUUCUUGAAGACCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAU ...(((((..(((((.((((......((((((.(((((((((........))))))))).))))))......)))).)))))..)))))....... ( -38.20) >DroSec_CAF1 91487 96 + 1 GAGAUCGGAAUCUCCUUCGAAGGCUUGGAGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAAGCGCCGGUAAAAGAU ...(((((....(((.((((......((((((.(((((((((........))))))))).))))))......)))).)))....)))))....... ( -30.60) >DroSim_CAF1 88575 96 + 1 GAGAUCGGGAUCUCUUUCGAAGGCUUGGAGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUGAGAGAU (((((....)))))(((((..(((..((((.((.....)).))))......(((((((.(((((......))))).))))))))))..)))))... ( -34.50) >DroEre_CAF1 88864 96 + 1 GUGAUCGGGGUCUCCUUUGAAGGCUUUGAGCUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAU ...(((((.(((((((((((((.....(((((.(((((((((........))))))))).)))))))))))).....)))))).)))))....... ( -39.30) >DroYak_CAF1 84181 96 + 1 GAGACCGGGAUCUCCUUUGCAGGCUUCGGGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAU ...(((((...(.((((..(..((((.(((((.(((((((((........))))))))).)))))....))))..)..)))).))))))....... ( -33.50) >DroAna_CAF1 78194 96 + 1 GCGACUUGCUUCUCUCCACUGGACUUGGAGUUCUUGAAGAUCCUCUUGAAGGACUUCAGCAGCUUCUUCAUGUUGGCGGAGGCACCAGUGAGAGAU ((.....)).((((((.(((((..(((((((((((.(((.....))).)))))))))))..((((((.(......).)))))).))))))))))). ( -39.80) >consensus GAGAUCGGGAUCUCCUUCGAAGGCUUGGAGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAU ...(((((..(((((......(((((((((((.(((((((((........))))))))).)))))...))))))...)))))..)))))....... (-30.26 = -29.98 + -0.27)
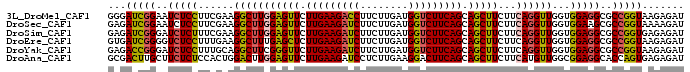
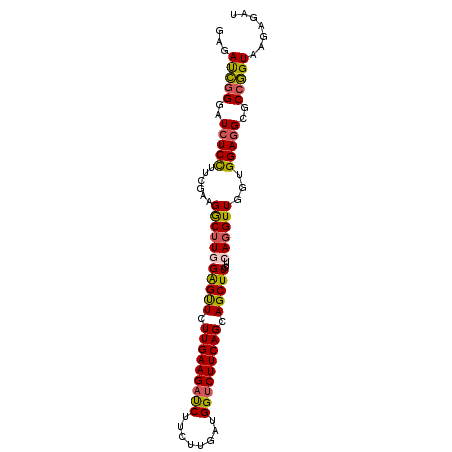
| Location | 14,909,418 – 14,909,532 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -29.13 |
| Energy contribution | -29.33 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14909418 114 + 23771897 UCGAAGGCUUGGAGUUCUUGAAGACCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAUUUCUUCUCUGUCACUCCA---UUGAAGUUGGAAA---CAA ..........((((((.(((((((((........))))))))).))))))(((((.(..((((((..((.....(((((.....)))))))..)))))---)..)..)))))..---... ( -39.10) >DroSec_CAF1 91503 114 + 1 UCGAAGGCUUGGAGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAAGCGCCGGUAAAAGAUUUCUUCUCUGGCAUUCCA---UUGAAGUUGGAAA---CUA ..........((((((.(((((((((........))))))))).))))))(((((.(..((((((..(((((...(((....)))..))))).)))))---)..)..)))))..---... ( -39.90) >DroSim_CAF1 88591 114 + 1 UCGAAGGCUUGGAGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUGAGAGAUUUCUUCUCUGGCAUUCCA---UUGAAGUUGGAAA---CUA ..........((((((.(((((((((........))))))))).))))))(((((.(..((((((..(((((.((.((....)))).))))).)))))---)..)..)))))..---... ( -40.80) >DroYak_CAF1 84197 114 + 1 UUGCAGGCUUCGGGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAUUUCUUCUCUGGCACUCCA---UUGAAGUUGGAAA---CGA .....(((((((((((.(((((((((........))))))))).))))).........(((((((..(((((.(((......)))..))))).)))))---))))))))(....---).. ( -40.60) >DroAna_CAF1 78210 117 + 1 CACUGGACUUGGAGUUCUUGAAGAUCCUCUUGAAGGACUUCAGCAGCUUCUUCAUGUUGGCGGAGGCACCAGUGAGAGAUUGUUUCUCAGAAACUCCG---UUGGAGUUGGCGGUGGAAA ((((((..(((((((((((.(((.....))).)))))))))))..((((((.(......).)))))).)))))).((((.....)))).....(.(((---((......))))).).... ( -43.30) >DroPer_CAF1 135625 117 + 1 UGCCCGCCUUCGAGUGGGUGAAGAUCCGCCUGAUGCUCUUUAGCAGCUUCUUCAUGUUGGUGGCGGCGCCGGUGAGAGAGCUCUUCUCCGGCACACCAGAGCUGGAGUUGAAAC---GAU .(((.(((...(((((((((......)))))...)))).....((((........))))..))))))(((((.((((....))))..)))))((.(((....))).))......---... ( -39.50) >consensus UCGAAGGCUUGGAGUUCUUGAAGAUCUUCUUGAUGGUCUUCAGCAGCUUCUUCAGGUUGGUGGAGGCGCCGGUAAGAGAUUUCUUCUCUGGCACUCCA___UUGAAGUUGGAAA___CAA ...........(((((.(((((((((........))))))))).)))))((((((.....(((((..(((((...(((....)))..))))).)))))...))))))............. (-29.13 = -29.33 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:27 2006