| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,905,602 – 14,905,716 |
| Length | 114 |
| Max. P | 0.610077 |

| Location | 14,905,602 – 14,905,716 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -25.73 |
| Energy contribution | -26.35 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
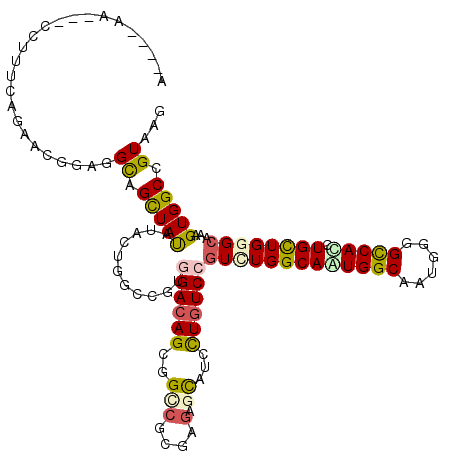
>3L_DroMel_CAF1 14905602 114 + 23771897 -----AAUUCCCCGGCAGAACGGAGGUAGCUACAUACUGGCUGUGGACAGUGGACGCGAGAGCAUUCUGUCCGUGUGGCAGUGGCAAUGGGGUCACCUGCUGGGCAAAGUGGCCGUAAG -----.....((((((((..........(((((((((.....))((((((.....((....))...))))))))))))).(((((......)))))))))))))............... ( -42.70) >DroVir_CAF1 91235 115 + 1 AAAACA----CUUUUCAGAACGGCGGCAGCUAUAUUCUUGCCGUGGACAGCGGCCGGGAGAGCAUCCUAUCCGUUUGGCAGUGGCAAUGGGGCCACCUGUUGGGCAAAGUGGCCGUAAG ......----.........(((((.((.(((...((((.(((((.....))))).)))).))).........(((..((((((((......)))).))))..)))...)).)))))... ( -46.50) >DroGri_CAF1 85170 114 + 1 AAACUAA-----UUUUAGAAUGGCGGCAGUUAUAUACUUGCCGUGGACAGUGGACGUGAGAGCAUUUUGUCCGUCUGGCAGUGGCAGUGGGGCCAUCUGCUGGGCAAGGUGGCAGUAAG ..(((.(-----((((......((((((((.....)).))))))((((((((..(....)..)))..)))))(((..((((((((......)))).))))..))))))))...)))... ( -41.60) >DroWil_CAF1 461930 109 + 1 ------A----UUUCCAGAACGGAGGCAGUUAUAUUCUUGCUGUUGACAGCGGCCGGGAGAGCAUAUUGUCUGUUUGGCAAUGGCAAUGGGGUCACCUGCUGGGCAAAGUGGCUGUAUG ------.----.((((.....))))((((((((.....(((((....)))))(((.((...((.(((((((.....)))))))))...(((....))).)).)))...))))))))... ( -37.00) >DroMoj_CAF1 116789 119 + 1 AAUCCAAUUGCCUUUCAGAAUGGAGGCAGCUAUAUACUGGCCGUGGACAGCGGUCGCGAGAGCAUCCUGUCCGUCUGGCAAUGGCAAUGGGGCCACCUGCUGGGCAAAGUGGCCGUAAG .......((((((((......)))))))).....(((.(((((.((((((.(((.((....)))))))))))(((..(((.((((......))))..)))..)))....)))))))).. ( -51.80) >DroAna_CAF1 74690 115 + 1 A----AAUCCCCUUUCAGAAUGGCGGCAGCUACAUCCUGGCAGUGGACAGUGGCCGUGAAAGUAUCCUGUCGGUGUGGCAAUGGCAGUGGGGUCAUCUGCUGGGGAAAGUGGCCGUAAG .----.................(((((.(((...(((..((((..(((....(((((....((..((....))....)).)))))......)))..))))..)))..))).)))))... ( -37.30) >consensus A____AA___CCUUUCAGAACGGAGGCAGCUAUAUACUGGCCGUGGACAGCGGCCGCGAGAGCAUCCUGUCCGUCUGGCAAUGGCAAUGGGGCCACCUGCUGGGCAAAGUGGCCGUAAG .........................((.(((((...........((((((..(((....).))...))))))(((((((((((((......))))).))))))))...))))).))... (-25.73 = -26.35 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:25 2006