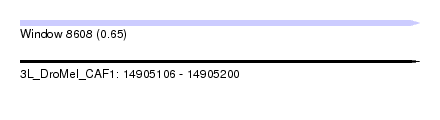
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,905,106 – 14,905,200 |
| Length | 94 |
| Max. P | 0.652616 |

| Location | 14,905,106 – 14,905,200 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.94 |
| Mean single sequence MFE | -37.34 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
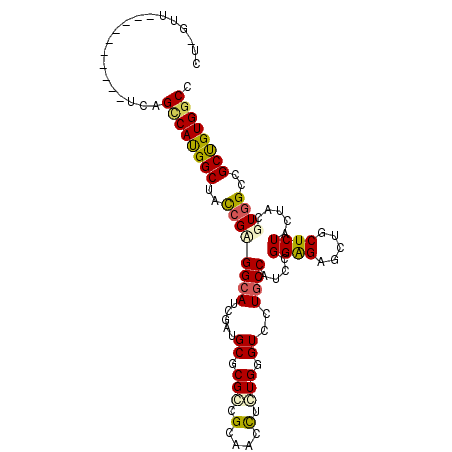
>3L_DroMel_CAF1 14905106 94 + 23771897 CU-GUU----------UUAGCCAUGGCUAUCGCGGCAUUGAUGCGCGGCGAAAUCUCUGGGUUUUGCCCUCCGGAGAACUGCUCUACUACGUGGCUGCCGUGGCC ..-...----------...((((((((..((((((((....)))..(((((((((....)))))))))....((((.....))))....)))))..)))))))). ( -39.10) >DroPse_CAF1 133005 95 + 1 CUCUUU----------UCAGCCAUGGCUACCGUGGCAUCGAUGCUCGCCGGAACCUUUGGGUCUUGCCGUCCGGGGAGCUGCUCUACUACGUGGCCGCUGUGGCC ......----------...(((((((((((.((((.......((((.((((((((....))).......))))).)))).......))))))))))...))))). ( -38.35) >DroGri_CAF1 84415 94 + 1 UU-AUU----------UUAGCCAUGGCUAUCGAGGCAUCGAUGCACGCCGCAACUUGUGGGUGCUGCCAACGGGAGAGCUCCUCUACUAUGUGGCAGCUGUGGCC ..-...----------...((((((((((((((....)))))((((.((((.....)))))))).((((...((((((...)))).))...))))))))))))). ( -40.90) >DroMoj_CAF1 116347 103 + 1 CU-GUUUGGCUUUCGA-UAGACACGGCUACAGAGGAAUCGAUGCGCGCCGCAAUCUCUGGGUACUGCCAAGCGGAGAGCUGCUCUACUACGUAGCUGCCGUGGCC ..-....(((..((..-..))((((((..((((((......((((...)))).))))))....((((...))))..((((((........))))))))))))))) ( -32.30) >DroAna_CAF1 74239 95 + 1 CUUUUU----------UCAGCCAUGGCUACCGCGGCAUUGAUGCCCGGCGUAAUCUCUGGGUUCUGCCCUCCGGAGAACUGCUCUACUAUGUGGCCGCUGUGGCC ......----------...((((((((..((((((((....)))).((((...((((((((........))))))))..)))).......))))..)))))))). ( -37.10) >DroPer_CAF1 131523 95 + 1 CUACUU----------UCAGUCAUGGCUACCGUGGCAUCGAUGCUCGCCGGAACCUGUGGGUCUUGCCGUCCGGGGAGCUGCUCUACUACGUGGCCGCUGUGGCC ......----------........((((((.(((((......((((.((((((((....))).......))))).)))).((........)).))))).)))))) ( -36.31) >consensus CU_GUU__________UCAGCCAUGGCUACCGAGGCAUCGAUGCGCGCCGCAACCUCUGGGUCCUGCCAUCCGGAGAGCUGCUCUACUACGUGGCCGCUGUGGCC ...................((((((((..((((((((.....((.(((.(....).))).))..))))....((((.....)))).....))))..)))))))). (-23.60 = -23.38 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:24 2006