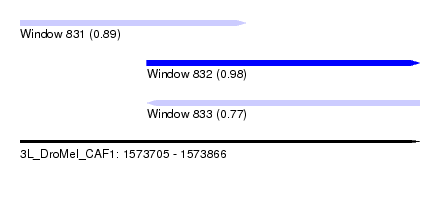
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,573,705 – 1,573,866 |
| Length | 161 |
| Max. P | 0.984879 |

| Location | 1,573,705 – 1,573,796 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -11.67 |
| Energy contribution | -14.87 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1573705 91 + 23771897 CA---AACUGCUGCCCACAUUCCUUGUGU--UUGUCU-CUGUUGAUUUGUUUCCAAUUUCCUACAACGAUACAAGCCAACAUAAGCAAGUAGCCCCU ..---....(((((.((((.....)))).--(((..(-.(((((..((((.((..............)).))))..))))).)..)))))))).... ( -19.04) >DroSec_CAF1 17996 93 + 1 CA---AACUGCUGCCCACAUUCCUUGUGUCUUUGGCU-UUGCUGGUUUGUUUCCAAUUUCCUACAACGCUACAAGCCAACAUGAGCAAGUAGCUCAU ..---....(((((.((((.....))))......(((-(((.((((((((....................)))))))).)).))))..))))).... ( -22.95) >DroSim_CAF1 18365 81 + 1 CA---AACUGCUGCCCACAUUCCUUGUGU-------------UGGUUUGUUUCCAAUUUCCUACAACGCUACAAGCCAACACGACCAAGUAGCUCCU ..---....(((((.........((((((-------------((((((((....................))))))))))))))....))))).... ( -21.57) >DroEre_CAF1 24968 94 + 1 UUGG-ACCUGCUGCCCACAUUCCUGGUGU--UUGGCUUUUGUUGGUUUCUUGCCAAUUUCCUACAACGAUACAAGCCAACAUGAGCUAGUAACCCCU .(((-.........))).......(((..--(((((((.(((((((((.(((..............)))...))))))))).)))))))..)))... ( -20.84) >DroYak_CAF1 19044 92 + 1 CAGAUAUCUGCUGCCCACAUUCCUUGUAC--UUCGCU-UUGUUGGUUUGUUGCCAAUUUCCUAGAACGCUAGAUGCCAACAUGAGCGAGUAUCCC-- (((....)))...............((((--(.((((-(((((((((((....)))....((((....))))..))))))).))))))))))...-- ( -22.10) >consensus CA___AACUGCUGCCCACAUUCCUUGUGU__UUGGCU_UUGUUGGUUUGUUUCCAAUUUCCUACAACGCUACAAGCCAACAUGAGCAAGUAGCCCCU .........(((((.((((.....))))......(((..(((((((((((....................)))))))))))..)))..))))).... (-11.67 = -14.87 + 3.20)

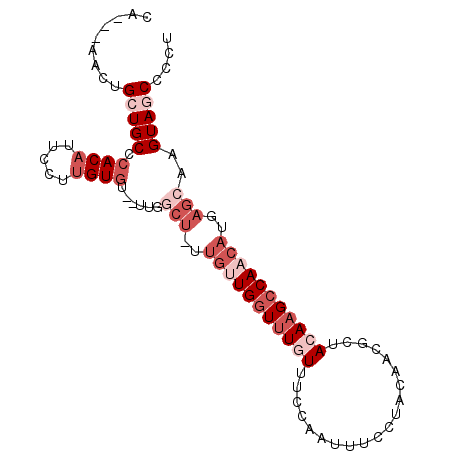
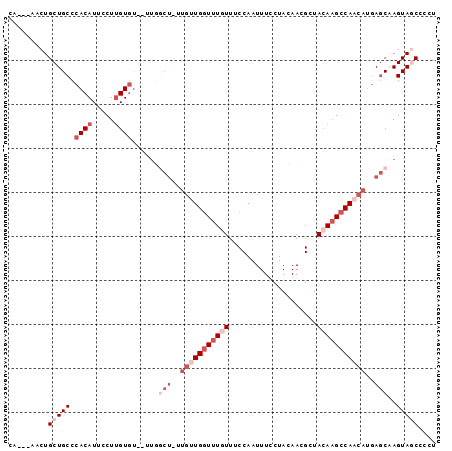
| Location | 1,573,756 – 1,573,866 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1573756 110 + 23771897 UUCCUACAACGAUACAAGCCAACAUAAGCAAGUAGCCCCUCCACCGACCGAUAGCCUCCAUGGCUUACAUGCUCACAUUGGCGUUGGCUUGGGAUCGUCA------AUCGUCAUCA ........(((((.(((((((((....((.....)).................(((....((((......)).))....))))))))))))..)))))..------.......... ( -29.40) >DroSec_CAF1 18049 105 + 1 UUCCUACAACGCUACAAGCCAACAUGAGCAAGUAGCUCAUCCAC----CGAUAGCCAUCAUAGCUUACAUGAUCACAUUGGCGUUGGUUUGGGGUCGUCA------AUCGUCAU-C ........((((..(((((((((((((((.....))))))...(----((((.(..(((((.......)))))..)))))).)))))))))..).)))..------........-. ( -30.10) >DroSim_CAF1 18406 106 + 1 UUCCUACAACGCUACAAGCCAACACGACCAAGUAGCUCCUCCAC----CGAUAGCCACCAUAGCUCACAUGCUCACAUUGGCGUUGGCUUGGGGUCGUCA------AUCGUCAUCC ........((((..(((((((((((......))...........----.....((((....(((......))).....)))))))))))))..).)))..------.......... ( -28.10) >DroEre_CAF1 25022 108 + 1 UUCCUACAACGAUACAAGCCAACAUGAGCUAGUAACCCCUCCUG----CU-CAGC--CCAAAGCUUACAAGCUCACAUUGGCGUUGGCUUGGGGUCGUCAUCGAGAGUAGUCAU-C ...((((..((((.(((((((((.(((((..(........)..)----))-))((--(...((((....))))......))))))))))))..))))((.....))))))....-. ( -35.60) >DroYak_CAF1 19098 95 + 1 UUCCUAGAACGCUAGAUGCCAACAUGAGCGAGUAUCCC--------------------CAUGGCUUACAAGCUCACAUUGGCGUUGGCUUGGGAUCGUCAUCUAGCGUCGUCAU-C ......((.(((((((((((((..(((((..(((..((--------------------...))..)))..)))))..)))))...(((........))))))))))))).....-. ( -34.00) >consensus UUCCUACAACGCUACAAGCCAACAUGAGCAAGUAGCCCCUCCAC____CGAUAGCC_CCAUAGCUUACAUGCUCACAUUGGCGUUGGCUUGGGGUCGUCA______AUCGUCAU_C .(((((....((((...(((((..((((((.((((((........................)))).)).))))))..)))))..)))).)))))...................... (-17.66 = -18.70 + 1.04)

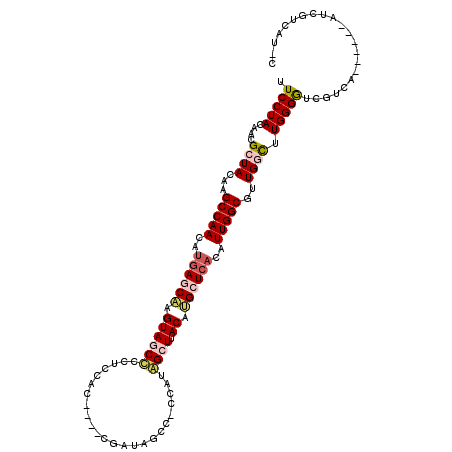
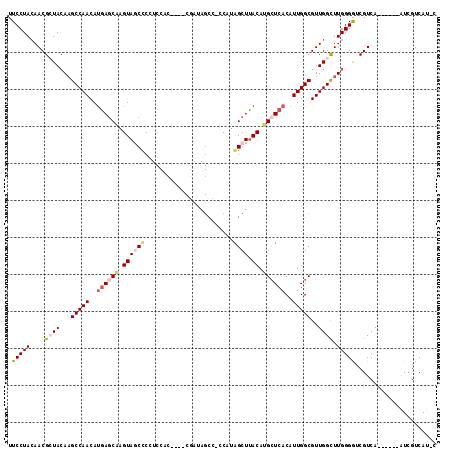
| Location | 1,573,756 – 1,573,866 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1573756 110 - 23771897 UGAUGACGAU------UGACGAUCCCAAGCCAACGCCAAUGUGAGCAUGUAAGCCAUGGAGGCUAUCGGUCGGUGGAGGGGCUACUUGCUUAUGUUGGCUUGUAUCGUUGUAGGAA ..........------.((((((..(((((((((......(((((((.(((.(((.......(((((....)))))...)))))).)))))))))))))))).))))))....... ( -37.70) >DroSec_CAF1 18049 105 - 1 G-AUGACGAU------UGACGACCCCAAACCAACGCCAAUGUGAUCAUGUAAGCUAUGAUGGCUAUCG----GUGGAUGAGCUACUUGCUCAUGUUGGCUUGUAGCGUUGUAGGAA .-........------.((((.(..(((.((((((((.((((.((((((.....)))))).)).)).)----))..((((((.....))))))))))).)))..)))))....... ( -31.40) >DroSim_CAF1 18406 106 - 1 GGAUGACGAU------UGACGACCCCAAGCCAACGCCAAUGUGAGCAUGUGAGCUAUGGUGGCUAUCG----GUGGAGGAGCUACUUGGUCGUGUUGGCUUGUAGCGUUGUAGGAA ..........------.((((.(..(((((((((((..(((....)))..((.(...(((((((.((.----...))..))))))).).)))))))))))))..)))))....... ( -36.10) >DroEre_CAF1 25022 108 - 1 G-AUGACUACUCUCGAUGACGACCCCAAGCCAACGCCAAUGUGAGCUUGUAAGCUUUGG--GCUG-AG----CAGGAGGGGUUACUAGCUCAUGUUGGCUUGUAUCGUUGUAGGAA .-....((((...(((((.......((((((((((((.....(((((....)))))..)--))((-((----(...((......)).))))).))))))))))))))..))))... ( -38.61) >DroYak_CAF1 19098 95 - 1 G-AUGACGACGCUAGAUGACGAUCCCAAGCCAACGCCAAUGUGAGCUUGUAAGCCAUG--------------------GGGAUACUCGCUCAUGUUGGCAUCUAGCGUUCUAGGAA .-.....((((((((((...(....)........((((((((((((..(((..((...--------------------.)).)))..))))))))))))))))))))))....... ( -35.90) >consensus G_AUGACGAU______UGACGACCCCAAGCCAACGCCAAUGUGAGCAUGUAAGCUAUGG_GGCUAUCG____GUGGAGGGGCUACUUGCUCAUGUUGGCUUGUAGCGUUGUAGGAA .................((((....((((((((((......((((((.(((.(((.......((((......))))...)))))).))))))))))))))))...))))....... (-22.82 = -23.50 + 0.68)


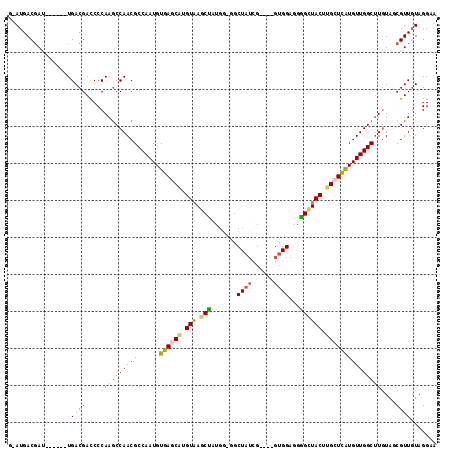
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:57 2006