| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,902,030 – 14,902,129 |
| Length | 99 |
| Max. P | 0.759141 |

| Location | 14,902,030 – 14,902,129 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
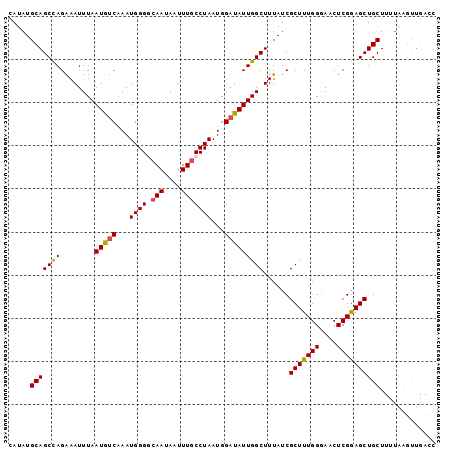
>3L_DroMel_CAF1 14902030 99 + 23771897 CAUAUGCAGCCAGAAAUUUAAUAUCAAAUGGGGCAAUAAUUUUCCUAAUGGAUAUUGGCUUUAUCGCUUUGGGAACUCGGAGCUGCUUUUAAGUUGACC .....(((((((........(((((...(((((.((....)))))))...)))))))))......(((((((....))))))))))............. ( -20.30) >DroSec_CAF1 84147 99 + 1 CAUAUGCAGCUAGAAAUUUAAUGUCAAAUGGGCCAAUAAUUUGCCUAAUGGAUAUUGGCUUUAUCGCUUUGGGAACUCGGAGCUGCUUUUAAGUUGACC ......(((((.((((.((((((((...(((((.((....)))))))...)))))))).......(((((((....)))))))...)))).)))))... ( -24.20) >DroSim_CAF1 80938 99 + 1 CAUAUGCAGCUAGAAAUUUAAUGUCAAAUGGGCCAAUAAUUUGCCUAAUGGAUAUUGGCUUUAUCGCUUUGGGAACUCGGAGCUGCUUUUAAGUUGACC ......(((((.((((.((((((((...(((((.((....)))))))...)))))))).......(((((((....)))))))...)))).)))))... ( -24.20) >DroEre_CAF1 81766 99 + 1 CAUAUGCAGCCAGAAAUUUAAUGUCAAAUGGGGCAAUAAUUUGCCUAAUGGAUAUUGGCUUUAUCGCUUUGGGAGCUCGGAGCUGCUUUUGAGUUGAAA ......(((((((((..((((((((.....((((((....))))))....)))))))).......(((((((....)))))))...))))).))))... ( -28.30) >DroYak_CAF1 76948 87 + 1 CAUAUGCAGCCAGGAAUUUAAUACCAAAUGGGGCAAUAAUUUGCCUAAUGGAUAUUGGCUUUAUCGCUUUGGAAACUCGAAGCUGCU------------ .....((((((((..(((((...((....))(((((....)))))...))))).)))))......(((((((....)))))))))).------------ ( -25.40) >consensus CAUAUGCAGCCAGAAAUUUAAUGUCAAAUGGGGCAAUAAUUUGCCUAAUGGAUAUUGGCUUUAUCGCUUUGGGAACUCGGAGCUGCUUUUAAGUUGACC .....(((((((........(((((...((((.(((....)))))))...)))))))))......(((((((....))))))))))............. (-20.48 = -20.24 + -0.24)


| Location | 14,902,030 – 14,902,129 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
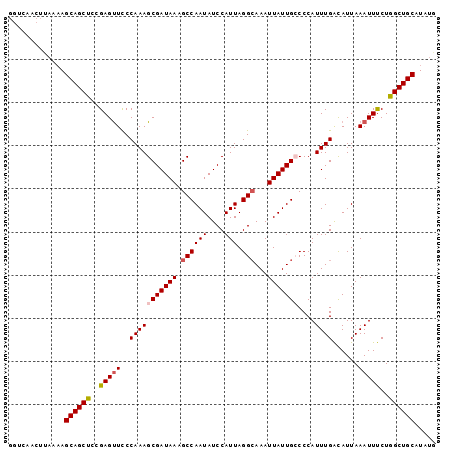
>3L_DroMel_CAF1 14902030 99 - 23771897 GGUCAACUUAAAAGCAGCUCCGAGUUCCCAAAGCGAUAAAGCCAAUAUCCAUUAGGAAAAUUAUUGCCCCAUUUGAUAUUAAAUUUCUGGCUGCAUAUG .............((((((..(((((..(((((((((((..(((((....))).))....)))))))....))))......)))))..))))))..... ( -16.60) >DroSec_CAF1 84147 99 - 1 GGUCAACUUAAAAGCAGCUCCGAGUUCCCAAAGCGAUAAAGCCAAUAUCCAUUAGGCAAAUUAUUGGCCCAUUUGACAUUAAAUUUCUAGCUGCAUAUG .............((((((..(((((.......(((....(((((((.((....)).....)))))))....)))......)))))..))))))..... ( -21.52) >DroSim_CAF1 80938 99 - 1 GGUCAACUUAAAAGCAGCUCCGAGUUCCCAAAGCGAUAAAGCCAAUAUCCAUUAGGCAAAUUAUUGGCCCAUUUGACAUUAAAUUUCUAGCUGCAUAUG .............((((((..(((((.......(((....(((((((.((....)).....)))))))....)))......)))))..))))))..... ( -21.52) >DroEre_CAF1 81766 99 - 1 UUUCAACUCAAAAGCAGCUCCGAGCUCCCAAAGCGAUAAAGCCAAUAUCCAUUAGGCAAAUUAUUGCCCCAUUUGACAUUAAAUUUCUGGCUGCAUAUG .............((((((..(((........(((((((.((((((....))).)))...)))))))...(((((....)))))))).))))))..... ( -17.80) >DroYak_CAF1 76948 87 - 1 ------------AGCAGCUUCGAGUUUCCAAAGCGAUAAAGCCAAUAUCCAUUAGGCAAAUUAUUGCCCCAUUUGGUAUUAAAUUCCUGGCUGCAUAUG ------------.((((((..((((((((((((((((((.((((((....))).)))...)))))))....)))))....))))))..))))))..... ( -26.50) >consensus GGUCAACUUAAAAGCAGCUCCGAGUUCCCAAAGCGAUAAAGCCAAUAUCCAUUAGGCAAAUUAUUGCCCCAUUUGACAUUAAAUUUCUGGCUGCAUAUG .............((((((..(((((..(((((((((((.((((((....))).)))...)))))))....))))......)))))..))))))..... (-16.36 = -16.76 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:23 2006