| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,890,648 – 14,890,751 |
| Length | 103 |
| Max. P | 0.835481 |

| Location | 14,890,648 – 14,890,751 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -5.10 |
| Energy contribution | -7.02 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.18 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14890648 103 + 23771897 AAACUGUUUGCAAAAUGAAUUUAUUAGUUAUGAGAUCA------CAAGGUCUGUGAAAGGCUCUUAAGACAACUAUCUGC-CAUAUAGGCUGUUCAACUUAAUUGCACAC---- .....((.(((((..(((((......(((.((((((((------((.....))))).....))))).)))........((-(.....))).)))))......))))).))---- ( -18.30) >DroSec_CAF1 72593 105 + 1 AAACGGUUUGCAAAUUGAAUUAAUUAGUUAUGAGAUCACGCCCACAAGGGCUGUGAAAGGCUCUGAAGACAAC----UGC-CAAAUAGGCUGUUCAACUUAAUUGCACAC---- .....((.(((((.((((........(((..(((.((((((((....)))).))))....)))....)))(((----.((-(.....))).)))....))))))))).))---- ( -25.70) >DroSim_CAF1 68590 105 + 1 AAACGGUUUGCAAAUUGAAUUAAUUAGUUAUGAGAUCACGCCCACAAGGGCUGUGAAAGGCUCUGAAGACAAC----UGC-CAAAUAGGCUGUUCAACUUAAUUGCACAC---- .....((.(((((.((((........(((..(((.((((((((....)))).))))....)))....)))(((----.((-(.....))).)))....))))))))).))---- ( -25.70) >DroEre_CAF1 69203 105 + 1 AAACUGUGUGCAAAAUGAAUCAGUUAG---UGAGAUAACACUCACAAGU-----GCAUGGCUAUUGAGACAAC-AUCUGCCCAAGUGGGCUGUUCAACCUAAUUGCACACCAGU ..(((((((((((..((..(((((..(---((((......))))).(((-----.....))))))))..))..-....((((....))))............))))))).)))) ( -30.50) >DroYak_CAF1 65126 110 + 1 AAACUGUGUGCAAAAUGAAUUAAUUAGUUAUGAGAUAGCACUCACAAGUGCUGUGCAUGACUAUUAAGACAACUGUCUGCACAAGUAGGCUGUUCAACUUAAGUGCACAC---- .....(((((((............((((((((..((((((((....)))))))).))))))))(((((..(((.((((((....)))))).)))...))))).)))))))---- ( -39.70) >consensus AAACUGUUUGCAAAAUGAAUUAAUUAGUUAUGAGAUCACACCCACAAGGGCUGUGAAAGGCUCUUAAGACAAC__UCUGC_CAAAUAGGCUGUUCAACUUAAUUGCACAC____ .....((.(((((.............(((.(((((.......((((.....))))......))))).)))...............((((........)))).))))).)).... ( -5.10 = -7.02 + 1.92)

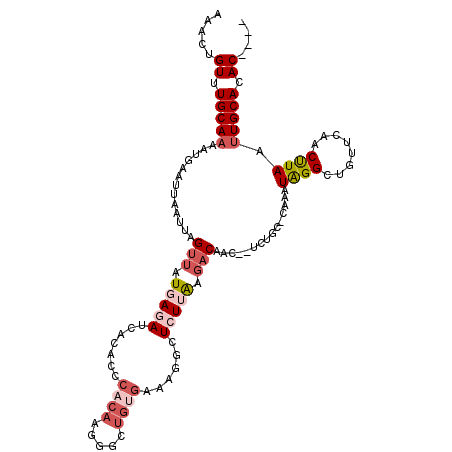
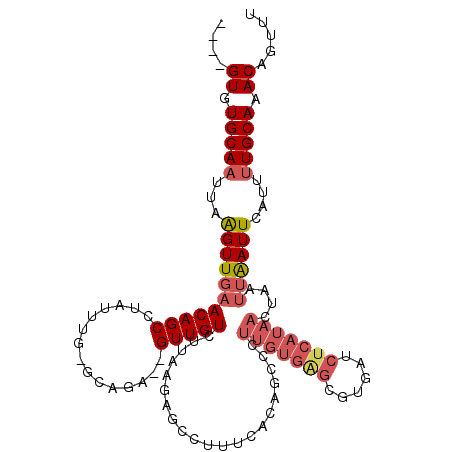
| Location | 14,890,648 – 14,890,751 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -9.22 |
| Energy contribution | -10.74 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14890648 103 - 23771897 ----GUGUGCAAUUAAGUUGAACAGCCUAUAUG-GCAGAUAGUUGUCUUAAGAGCCUUUCACAGACCUUG------UGAUCUCAUAACUAAUAAAUUCAUUUUGCAAACAGUUU ----((.(((((......((((..(((.....)-))...(((((((.....(((....((((((...)))------))).)))))))))).....))))..))))).))..... ( -21.90) >DroSec_CAF1 72593 105 - 1 ----GUGUGCAAUUAAGUUGAACAGCCUAUUUG-GCA----GUUGUCUUCAGAGCCUUUCACAGCCCUUGUGGGCGUGAUCUCAUAACUAAUUAAUUCAAUUUGCAAACCGUUU ----((.(((((....((((((..(((.....)-))(----(((((.....(((....((((.((((....)))))))).)))))))))......))))))))))).))..... ( -30.10) >DroSim_CAF1 68590 105 - 1 ----GUGUGCAAUUAAGUUGAACAGCCUAUUUG-GCA----GUUGUCUUCAGAGCCUUUCACAGCCCUUGUGGGCGUGAUCUCAUAACUAAUUAAUUCAAUUUGCAAACCGUUU ----((.(((((....((((((..(((.....)-))(----(((((.....(((....((((.((((....)))))))).)))))))))......))))))))))).))..... ( -30.10) >DroEre_CAF1 69203 105 - 1 ACUGGUGUGCAAUUAGGUUGAACAGCCCACUUGGGCAGAU-GUUGUCUCAAUAGCCAUGC-----ACUUGUGAGUGUUAUCUCA---CUAACUGAUUCAUUUUGCACACAGUUU ((((.(((((((...(((((....((((....))))((((-...))))...)))))....-----....(((((......))))---).............))))))))))).. ( -32.00) >DroYak_CAF1 65126 110 - 1 ----GUGUGCACUUAAGUUGAACAGCCUACUUGUGCAGACAGUUGUCUUAAUAGUCAUGCACAGCACUUGUGAGUGCUAUCUCAUAACUAAUUAAUUCAUUUUGCACACAGUUU ----(((((((....((((((...........((((((((.((((...)))).))).)))))((((((....)))))).............)))))).....)))))))..... ( -30.40) >consensus ____GUGUGCAAUUAAGUUGAACAGCCUAUUUG_GCAGA__GUUGUCUUAAGAGCCUUUCACAGCCCUUGUGAGCGUGAUCUCAUAACUAAUUAAUUCAUUUUGCAAACAGUUU ....((.(((((...(((((((((((...............))))).....................(((((((......)))))))....))))))....))))).))..... ( -9.22 = -10.74 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:14 2006