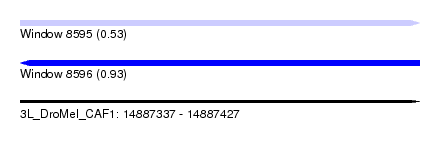
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,887,337 – 14,887,427 |
| Length | 90 |
| Max. P | 0.928951 |

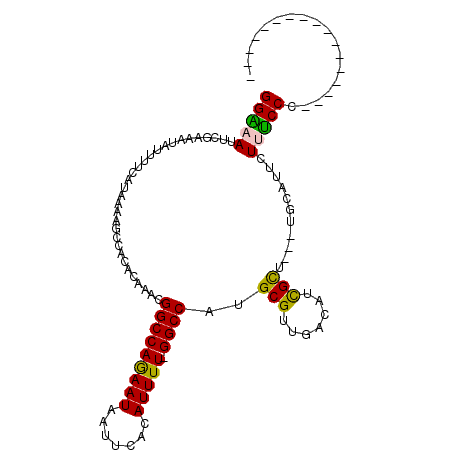
| Location | 14,887,337 – 14,887,427 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -16.73 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14887337 90 + 23771897 -----------------GGGAAAAAAUGCA---UGCGAUGUCAUCGCAUGGCC-AAAAUGUGAAUUAUUCUGGCCGUUCUUGUGGCUUUUAUGAAAAUAUUUCGAAUUUCC -----------------.(((((.....((---((((((...))))))))..(-.((((((...((((...(((((......)))))...))))..)))))).)..))))) ( -23.00) >DroPse_CAF1 117505 108 + 1 AGGUGCCCGGGGAGUUUGGGGGAGUCUCUG---AACGAUGUCAACGCAUGGCCGAAAAUGUGAAUUAUUCUGGCCGGUUGUGUGGCUUUUAUGAAAAUAUUUCAAAUUCCC .........(((((((((..((((((..((---(......)))((((((((((((((((....))).))).)))))..)))))))))).(((....))))..))))))))) ( -31.30) >DroSec_CAF1 69534 90 + 1 -----------------GGGAAAAAAUGCA---UGCGAUGUCAUCGCAUGGCC-AAAAUGUGAAUUAUUCUGGCCGUUCAUGUGGCUUUUGUGAAAAUAUUUCGAAUUUCC -----------------.(((((.....((---((((((...))))))))..(-.((((((...((((...(((((......)))))...))))..)))))).)..))))) ( -23.30) >DroEre_CAF1 65814 89 + 1 -----------------GGGAAAAAAUGCA---AGCGAUGUCAACGCAUGGCC-AAAAUGUGAAUUAUUCUGGCCGUUC-UGUGGCUUUUAUGAAAAUAUUUCGAAUUUCC -----------------.(((((..(((.(---((((....)..(((((((((-(.((((.....)))).))))))...-)))))))).)))(((.....)))...))))) ( -20.00) >DroAna_CAF1 57683 94 + 1 -----------------GGGAAAGGGAGAAAGGGGCAAUGUCAACGCAUGGCCGAAAAUGUGAAUUAUUUUGGCCGAUUGUGUGGCUUUUAUGAAAAUACUUCGAAUUCCC -----------------(((((..((((...........((((.(((((((((.((((((.....)))))))))))..))))))))...(((....)))))))...))))) ( -26.60) >DroPer_CAF1 115835 108 + 1 AGGUGCCUGGGGAGUUUGGGGGAGUCUCUG---AACGAUGUCAACGCAUGGCCGAAAAUGUGAAUUAUUCUGGCCGGUUGUGUGGCUUUUAUGAAAAUAUUUCAAAUUCCC .........(((((((((..((((((..((---(......)))((((((((((((((((....))).))).)))))..)))))))))).(((....))))..))))))))) ( -31.30) >consensus _________________GGGAAAAAAUGCA___AGCGAUGUCAACGCAUGGCC_AAAAUGUGAAUUAUUCUGGCCGUUCGUGUGGCUUUUAUGAAAAUAUUUCGAAUUCCC .................(((((............((.((((....)))).))...((((((...((((...(((((......)))))...))))..))))))....))))) (-16.73 = -16.40 + -0.33)

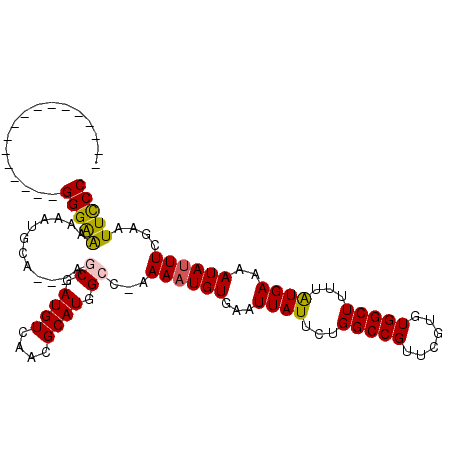
| Location | 14,887,337 – 14,887,427 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14887337 90 - 23771897 GGAAAUUCGAAAUAUUUUCAUAAAAGCCACAAGAACGGCCAGAAUAAUUCACAUUUU-GGCCAUGCGAUGACAUCGCA---UGCAUUUUUUCCC----------------- (((((...(((.....)))......((.........(((((((((.......)))))-))))(((((((...))))))---)))....))))).----------------- ( -24.70) >DroPse_CAF1 117505 108 - 1 GGGAAUUUGAAAUAUUUUCAUAAAAGCCACACAACCGGCCAGAAUAAUUCACAUUUUCGGCCAUGCGUUGACAUCGUU---CAGAGACUCCCCCAAACUCCCCGGGCACCU ((((.((((......((((.................(((((((((.......))))).))))..(((.......))).---..))))......)))).))))......... ( -19.10) >DroSec_CAF1 69534 90 - 1 GGAAAUUCGAAAUAUUUUCACAAAAGCCACAUGAACGGCCAGAAUAAUUCACAUUUU-GGCCAUGCGAUGACAUCGCA---UGCAUUUUUUCCC----------------- (((((...(((.....)))......((.........(((((((((.......)))))-))))(((((((...))))))---)))....))))).----------------- ( -24.70) >DroEre_CAF1 65814 89 - 1 GGAAAUUCGAAAUAUUUUCAUAAAAGCCACA-GAACGGCCAGAAUAAUUCACAUUUU-GGCCAUGCGUUGACAUCGCU---UGCAUUUUUUCCC----------------- (((((...(((.....)))......((....-....(((((((((.......)))))-))))..(((.......))).---.))....))))).----------------- ( -20.50) >DroAna_CAF1 57683 94 - 1 GGGAAUUCGAAGUAUUUUCAUAAAAGCCACACAAUCGGCCAAAAUAAUUCACAUUUUCGGCCAUGCGUUGACAUUGCCCCUUUCUCCCUUUCCC----------------- ((((....(((.((((((.......(((........))).)))))).)))........(((.(((......))).)))......))))......----------------- ( -16.90) >DroPer_CAF1 115835 108 - 1 GGGAAUUUGAAAUAUUUUCAUAAAAGCCACACAACCGGCCAGAAUAAUUCACAUUUUCGGCCAUGCGUUGACAUCGUU---CAGAGACUCCCCCAAACUCCCCAGGCACCU ((((.((((......((((.................(((((((((.......))))).))))..(((.......))).---..))))......)))).))))......... ( -19.10) >consensus GGAAAUUCGAAAUAUUUUCAUAAAAGCCACACAAACGGCCAGAAUAAUUCACAUUUU_GGCCAUGCGUUGACAUCGCU___UGCAUUCUUUCCC_________________ (((((...............................(((((((((.......))))).))))..(((.......)))...........))))).................. (-14.69 = -14.05 + -0.64)

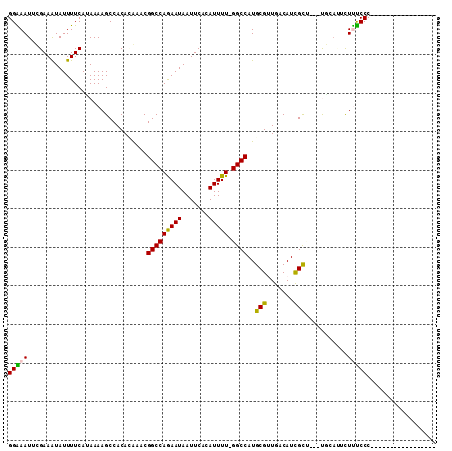
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:12 2006