
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,877,662 – 14,877,798 |
| Length | 136 |
| Max. P | 0.798124 |

| Location | 14,877,662 – 14,877,759 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
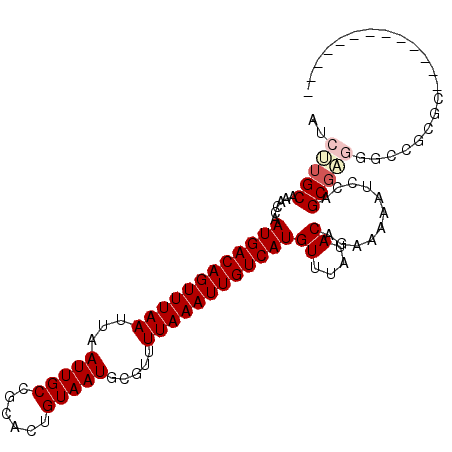
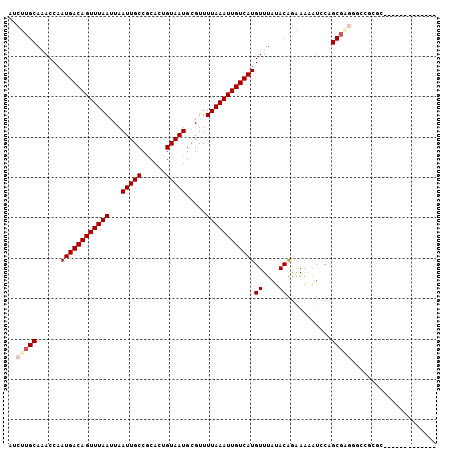
>3L_DroMel_CAF1 14877662 97 - 23771897 AUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAAAUCCAGCGAGUGCCUCUGUAUU--------- .......((((..((((((((((((...(((((......))))).....))))))))))))))))(((((((....((....)).....))))))).--------- ( -20.60) >DroPse_CAF1 107573 92 - 1 GUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAAAUGCUGCGAGGCUCGCC-------------- ((((((((.....((((((((((((...(((((......))))).....))))))))))))((....))..........)))))))).....-------------- ( -24.80) >DroGri_CAF1 62372 92 - 1 AUCGUGCA-ACCAAUGACAGUUUAAUUAAUUGCCACACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAACAGUGUCCGGCGGCUGUCGAGC------------- .(((.((.-.((.((((((((((((...(((((......))))).....))))))))))))....((((....))))..))..))...)))..------------- ( -23.60) >DroYak_CAF1 51734 97 - 1 AUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACGGAAAAAUGCAGCCCUGGCCGCUGUAUU--------- ..........((.((((((((((((...(((((......))))).....))))))))))))((....))))...((((((((.......))))))))--------- ( -23.70) >DroMoj_CAF1 78450 92 - 1 AUCCUGCA-ACCAAUGACAGUUUAAUUAAUUGCCACACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGGAAUGUCCGGCGGGUGCCAAGA------------- .(((((..-....((((((((((((...(((((......))))).....)))))))))))).......)))))......(((....)))....------------- ( -25.72) >DroAna_CAF1 48467 106 - 1 AUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAAAUGCUGCGAGGCUCUCGCUGUGAACGGUUCG ........((((.((((((((((((...(((((......))))).....))))))))))))(((((((((......))..(((((...)))))))))))))))).. ( -27.60) >consensus AUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAAAUCCAGCGAGGGCCGCGC_____________ ..(((((......((((((((((((...(((((......))))).....))))))))))))((....))...........)))))..................... (-17.70 = -18.37 + 0.67)

| Location | 14,877,679 – 14,877,798 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.28 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
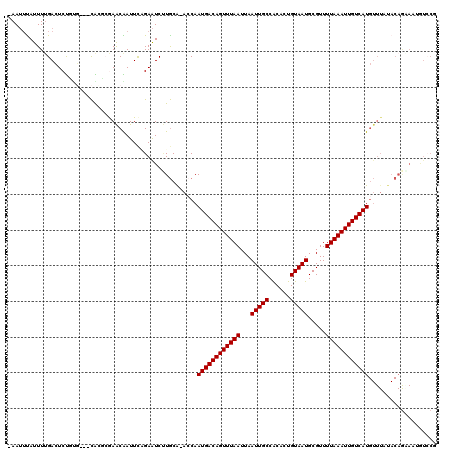
>3L_DroMel_CAF1 14877679 119 - 23771897 -AAUUUAUUUUGACUCUGUGGCGCGGCGAAACGAUUCAGAAUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAAAUCCA -.............((((((..((((.((..(......)..))))))((((..((((((((((((...(((((......))))).....)))))))))))))))).))))))........ ( -24.00) >DroVir_CAF1 67622 115 - 1 -AAUUUAUUUUGACUCUUUG---CACGCGAGCAAUUCUGAAUCCUGCG-AGCAAUGACAGUUUAAUUAAUUGCCACACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGCAAUGUCCG -..........(((...(((---(..((..(((...........))).-.)).((((((((((((...(((((......))))).....)))))))))))).........)))).))).. ( -27.00) >DroPse_CAF1 107585 120 - 1 GAAAGCAUUUUGACUCCGCCGAGCGGCGAAACGAUUCAGAGUCUUGCAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAAAUGCU ...(((((((((((((.(((....)))(((....))).)))))....((((..((((((((((((...(((((......))))).....)))))))))))))))).......)))))))) ( -34.00) >DroGri_CAF1 62385 115 - 1 -AAUUUAUUUGGACUCUUUG---CACACGAACAAUUCAGAAUCGUGCA-ACCAAUGACAGUUUAAUUAAUUGCCACACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAACAGUGUCCG -.........((((.(((((---(((.................)))))-)...((((((((((((...(((((......))))).....))))))))))))...........)).)))). ( -27.03) >DroWil_CAF1 370710 119 - 1 -AAUUUAUUUUGAAUCUGAUGUGUGCGCGAACGAUUCGGAAUCUUACAAACCAAUGACAGUUUAAUUAAUUGCCGCACUGUAAUGCGUUUUAAAUUGUCAUNNNNNNNNNNNNNNNNNNN -.....((((((((((...(((....)))...))))))))))...........((((((((((((...(((((......))))).....))))))))))))................... ( -22.30) >DroMoj_CAF1 78463 115 - 1 -AAUUUAUUUUGACUCUUUG---CACGCGAGCAAUUUCGAAUCCUGCA-ACCAAUGACAGUUUAAUUAAUUGCCACACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGGAAUGUCCG -.......(((((....(((---(......))))..)))))(((((..-....((((((((((((...(((((......))))).....)))))))))))).......)))))....... ( -26.62) >consensus _AAUUUAUUUUGACUCUGUG___CACGCGAACAAUUCAGAAUCUUGCA_ACCAAUGACAGUUUAAUUAAUUGCCACACUGUAAUGCGUUUUAAAUUGUCAUGUUUAUACAGAAAUGUCCG .....................................................((((((((((((...(((((......))))).....))))))))))))................... (-14.28 = -14.28 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:08 2006