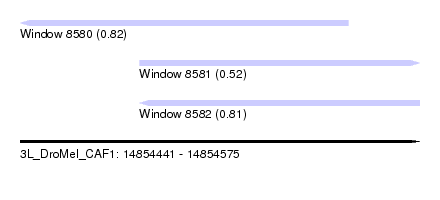
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,854,441 – 14,854,575 |
| Length | 134 |
| Max. P | 0.823448 |

| Location | 14,854,441 – 14,854,551 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -11.55 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14854441 110 - 23771897 UCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGUCCGGGUAUGUGG .((.((------(..(((.----((((.(((((((((((.....(((......)))))))))))))).............((((((((.......)))))))).)))).)))..))).)) ( -27.80) >DroVir_CAF1 49277 101 - 1 -------------AGUCGU----ACGAAAAUACUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAUGUAAUAAGCACUUUUAAUAAAUAAAGGUGAACUUUUA--AGCGAAUGAAG -------------..((((----(((..((((((((((.....((((......))))))))))))))..)))......((((((((.....))))))))........--.))))...... ( -24.50) >DroGri_CAF1 45123 103 - 1 -------------AGUCGU----UCGAAAAUGCUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGCUAAAGUAAUAAGCACUUUUAAUAAAUAAAGGUGAAUUGUUGUCGGAGAGUCAAA -------------..((.(----((((....(((((((.....((((......))))))))))).......((((((.((((((((.....))))))))..)))))))))))))...... ( -23.40) >DroWil_CAF1 285711 117 - 1 UCCUCAAUGCUUUUGUCGUGAGAACGAAAAU-CGUGAGAAAAUCGUGUUAAGUGGCGCGCAAGUGUUAAAAUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGCUGUAUGAGCGA-AA- ((((((.((((((((((....).)))))).(-(....))......(((((..((((((....))))))...)))))(((.((((((((.......)))))))))))))))))).))-..- ( -31.90) >DroMoj_CAF1 56186 100 - 1 -------------AGUCGU----AUAAAAAUACUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAUGUAAUAAGCAGUUUUAAUAAAUAAAGGUGA-CAUUUA--AGCGAAUGUUG -------------..((((----.((((((((((((((.....((((......))))))))))))))..........(((.(((((.....))))).)).-).))))--.))))...... ( -21.60) >DroPer_CAF1 83790 110 - 1 UCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAGAGGUUUCUUGGGUGUGAAA .(((((------((....(----((...(((((((((((.....(((......))))))))))))))...)))...)))(((((........)))))...))))((((........)))) ( -26.90) >consensus _____________AGUCGU____ACGAAAAUACUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGGUGAAAGGUUGU_AGAGAAUGAAG ...............(((......))).((((((((((.....((((......))))))))))))))...........((.(((((.....))))).))..................... (-11.55 = -12.30 + 0.75)

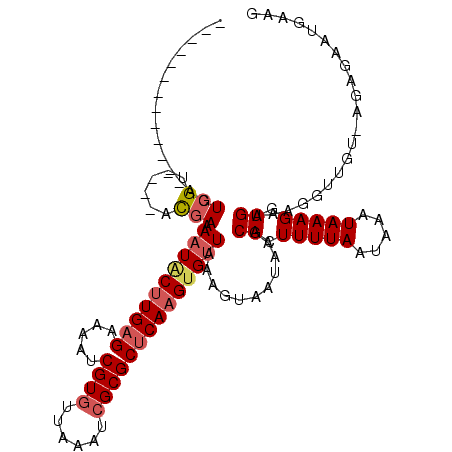
| Location | 14,854,481 – 14,854,575 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14854481 94 + 23771897 UGCUUAUUACUUUAACACUUGAGCGCGAUUUAACACGAUUUGCUCAAGUAUUUUCGUACGACAACGAGGACACAUUCCGGCGUUCUCGUUCUCG---------------- .......(((...((.(((((((((.((((......))))))))))))).))...)))(((.(((((((((.(......).))))))))).)))---------------- ( -27.60) >DroPse_CAF1 86349 85 + 1 UGCUUAUUACUUUAACACUUGAGCGCGAUUUAACACGAUUUGCUCAAGUAUUUUCGUACGACAACGAGGACACAUUCCUCCUUGA------------------------- .......(((...((.(((((((((.((((......))))))))))))).))...)))...(((.(((((.....))))).))).------------------------- ( -20.90) >DroSim_CAF1 31668 85 + 1 UGCUUAUUACUUUAACACUUGAGCGCGAUUUAACACGAUUUGCUCAGGUAUUUUCGUACGACAACGAGGACACAUUCCGGCGUUC------------------------- .(((............(((((((((.((((......))))))))))))).(((((((......)))))))........)))....------------------------- ( -19.20) >DroEre_CAF1 33081 97 + 1 UGCUUAUUACUUUAACACUUAAACGCGAUUUAACACGAUUUGCUCAAGCAUUUUCGUACGACAACGAGGACACAUUCCGGCGCUCCCCUUCUCCUCC------------- ........................(((.......((((..(((....)))...))))..........(((.....)))..)))..............------------- ( -13.00) >DroYak_CAF1 27661 110 + 1 UGCUUAUUACUUUAACACUUGAGCGCGAUUUAACACGAUUUGCUCAAGUAUUUUCGUACGACAACGAGGACACAUUCCGGCGUUCUCCUUCUCGUCCCCGUCCCCGUUCU .............((((((((((((.((((......)))))))))))))......(.((((.((.((((((.(......).)))))).)).)))).)........))).. ( -23.50) >DroPer_CAF1 83830 85 + 1 UGCUUAUUACUUUAACACUUGAGCGCGAUUUAACACGAUUUGCUCAAGUAUUUUCGUACGACAACGAGGACACAUUCCUCCUUGA------------------------- .......(((...((.(((((((((.((((......))))))))))))).))...)))...(((.(((((.....))))).))).------------------------- ( -20.90) >consensus UGCUUAUUACUUUAACACUUGAGCGCGAUUUAACACGAUUUGCUCAAGUAUUUUCGUACGACAACGAGGACACAUUCCGGCGUUC_________________________ ................(((((((((.((((......))))))))))))).(((((((......)))))))........................................ (-14.39 = -14.75 + 0.36)


| Location | 14,854,481 – 14,854,575 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14854481 94 - 23771897 ----------------CGAGAACGAGAACGCCGGAAUGUGUCCUCGUUGUCGUACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCA ----------------(((.((((((.((((......)))).)))))).)))(((...(((((((((((.....(((......))))))))))))))...)))....... ( -33.10) >DroPse_CAF1 86349 85 - 1 -------------------------UCAAGGAGGAAUGUGUCCUCGUUGUCGUACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCA -------------------------.(((.(((((.....))))).)))...(((...(((((((((((.....(((......))))))))))))))...)))....... ( -26.50) >DroSim_CAF1 31668 85 - 1 -------------------------GAACGCCGGAAUGUGUCCUCGUUGUCGUACGAAAAUACCUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCA -------------------------.((((..(((.....))).))))((..(((...(((((.(((((.....(((......)))))))).)))))...)))....)). ( -17.50) >DroEre_CAF1 33081 97 - 1 -------------GGAGGAGAAGGGGAGCGCCGGAAUGUGUCCUCGUUGUCGUACGAAAAUGCUUGAGCAAAUCGUGUUAAAUCGCGUUUAAGUGUUAAAGUAAUAAGCA -------------.........(((((.(((......))))))))(((....(((...(((((((((((.....(((......))))))))))))))...)))...))). ( -22.90) >DroYak_CAF1 27661 110 - 1 AGAACGGGGACGGGGACGAGAAGGAGAACGCCGGAAUGUGUCCUCGUUGUCGUACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCA ...((((.((((((((((....((......))......)))))))))).))))((...(((((((((((.....(((......))))))))))))))...))........ ( -38.50) >DroPer_CAF1 83830 85 - 1 -------------------------UCAAGGAGGAAUGUGUCCUCGUUGUCGUACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCA -------------------------.(((.(((((.....))))).)))...(((...(((((((((((.....(((......))))))))))))))...)))....... ( -26.50) >consensus _________________________GAACGCCGGAAUGUGUCCUCGUUGUCGUACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCA ................................(((.....)))..(((....(((...(((((((((((.....(((......))))))))))))))...)))...))). (-20.28 = -20.03 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:59 2006