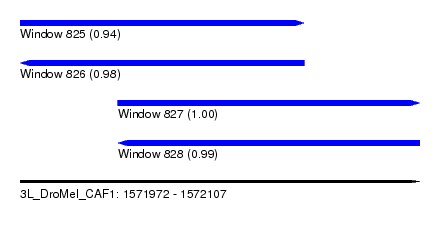
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,571,972 – 1,572,107 |
| Length | 135 |
| Max. P | 0.998543 |

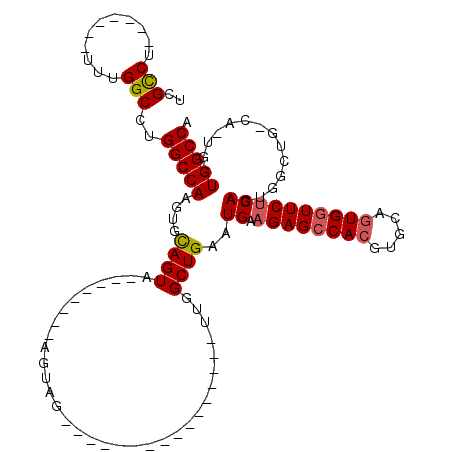
| Location | 1,571,972 – 1,572,068 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.78 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -22.53 |
| Energy contribution | -22.81 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1571972 96 + 23771897 UCGCCU-------UUUGGCCUGGCCAAGUGUAGUAAGUAAGUAAGUAG----------------UUGGCUGAAUGAAGAGCCACGUGCAGUGGUUCUCAGUGGCUGCCAGUGGUGGCCA ..(((.-------...)))..((((((.((...((......))..)).----------------))))))...(((.(((((((.....)))))))))).((((..((...))..)))) ( -35.00) >DroSec_CAF1 16456 78 + 1 UCGCCU-------UUUGGCCUGGCCAAGUGCAGUA--------AGUAG----------------UUGGCUGAAUGAAGAGCCAAGUGCAGUGGUUCUCAC----------UGGUGGCCA ..(((.-------...)))..(((((.........--------.(((.----------------((((((........)))))).)))((((.....)))----------)..))))). ( -29.10) >DroSim_CAF1 16639 78 + 1 UCGCCU-------UUUGGCCUGGCCAAGUGCAGUA--------AGUAG----------------UUGGCUGAAUGUAGAGCCACGUGCAGUGGUUCUCAC----------UGGUGGCCA ......-------..(((((.((((((.(((....--------.))).----------------))))))...((.((((((((.....)))))))))).----------....))))) ( -33.20) >DroEre_CAF1 22754 101 + 1 UCGUCU-------UUUGGCCUGGCCAAGUGCAGUA--------AGUGGUUGUUUGGGCCGAAUGUUGGCUGAGUGAAGAGCCACGUGCAGUGCUUCUCAGUGGCUGCCA---GUGGCCA ..(((.-------.(((((..(((((((..(.(((--------.((((((.((((((((((...))))))...)))).)))))).))).)..))......)))))))))---).))).. ( -37.10) >DroYak_CAF1 17183 96 + 1 UCGCCUUUUAGCUUUUGGCCUGGCCAAGUGCAGUA--------AGUAGUU------------UGUUGGCUGAAUGAAGAGCCACGUGCAGUGGUUCCCAGUGGCUGUCA---GUGGCCA ..(((...(((((.((((...(((((...((((..--------......)------------)))))))).......(((((((.....))))))))))).)))))...---..))).. ( -32.70) >consensus UCGCCU_______UUUGGCCUGGCCAAGUGCAGUA________AGUAG________________UUGGCUGAAUGAAGAGCCACGUGCAGUGGUUCUCAGUGGCUG_CA_UGGUGGCCA ..(((...........)))..(((((....((((.................................))))..((.((((((((.....))))))))))..............))))). (-22.53 = -22.81 + 0.28)

| Location | 1,571,972 – 1,572,068 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.78 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -18.37 |
| Energy contribution | -18.61 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1571972 96 - 23771897 UGGCCACCACUGGCAGCCACUGAGAACCACUGCACGUGGCUCUUCAUUCAGCCAA----------------CUACUUACUUACUUACUACACUUGGCCAGGCCAAA-------AGGCGA ((((((....((((((((((((((.....)).)).)))))).........)))).----------------.....((......)).......)))))).(((...-------.))).. ( -24.30) >DroSec_CAF1 16456 78 - 1 UGGCCACCA----------GUGAGAACCACUGCACUUGGCUCUUCAUUCAGCCAA----------------CUACU--------UACUGCACUUGGCCAGGCCAAA-------AGGCGA ((((((.((----------(((.....)))))...((((((........))))))----------------.....--------.........)))))).(((...-------.))).. ( -25.90) >DroSim_CAF1 16639 78 - 1 UGGCCACCA----------GUGAGAACCACUGCACGUGGCUCUACAUUCAGCCAA----------------CUACU--------UACUGCACUUGGCCAGGCCAAA-------AGGCGA ((((((.((----------(((((....((.....))((((........))))..----------------...))--------)))))....)))))).(((...-------.))).. ( -26.10) >DroEre_CAF1 22754 101 - 1 UGGCCAC---UGGCAGCCACUGAGAAGCACUGCACGUGGCUCUUCACUCAGCCAACAUUCGGCCCAAACAACCACU--------UACUGCACUUGGCCAGGCCAAA-------AGACGA (((((..---((((((((((((((.....)).)).)))))).........))))......((((............--------..........)))).)))))..-------...... ( -28.75) >DroYak_CAF1 17183 96 - 1 UGGCCAC---UGACAGCCACUGGGAACCACUGCACGUGGCUCUUCAUUCAGCCAACA------------AACUACU--------UACUGCACUUGGCCAGGCCAAAAGCUAAAAGGCGA (((((..---(((.((((((((.(......).)).))))))..)))....(((((..------------.......--------........)))))..)))))...(((....))).. ( -26.43) >consensus UGGCCACCA_UG_CAGCCACUGAGAACCACUGCACGUGGCUCUUCAUUCAGCCAA________________CUACU________UACUGCACUUGGCCAGGCCAAA_______AGGCGA (((((...............(((((.((((.....)))).))).))....(((((.....................................)))))..)))))............... (-18.37 = -18.61 + 0.24)

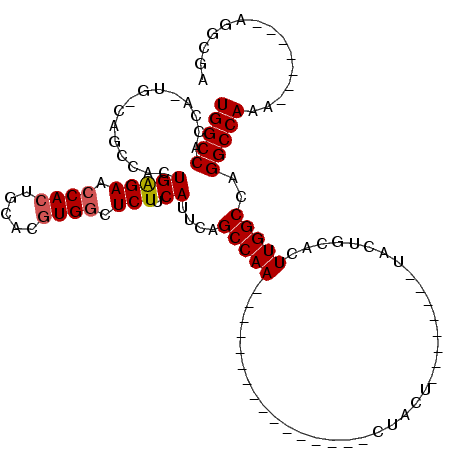
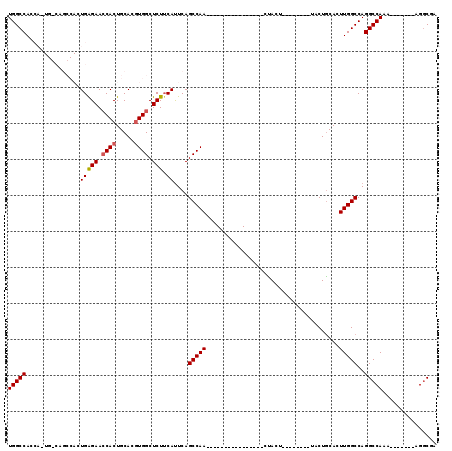
| Location | 1,572,005 – 1,572,107 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.74 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -22.73 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1572005 102 + 23771897 GUAAGUAG----------------UUGGCUGAAUGAAGAGCCACGUGCAGUGGUUCUCAGUGGCUGCCAGUGGUGGCCAACGAGCCAGCUAGCCACCCCAAACACCCAAUCCAUCCA-A .....(((----------------((((((...(((.(((((((.....)))))))))).((((..((...))..))))...)))))))))..........................-. ( -38.30) >DroSec_CAF1 16484 82 + 1 ---AGUAG----------------UUGGCUGAAUGAAGAGCCAAGUGCAGUGGUUCUCAC----------UGGUGGCCAACGAGCCGGCAAGCCACCCAAAACACCCAAUC-------- ---.(((.----------------((((((........)))))).))).((((((.....----------((.((((......)))).))))))))...............-------- ( -26.20) >DroSim_CAF1 16667 89 + 1 ---AGUAG----------------UUGGCUGAAUGUAGAGCCACGUGCAGUGGUUCUCAC----------UGGUGGCCAACGAGCCAGCAAGCCACCCAAAACACCCAAUCCAUCCA-A ---....(----------------((((.((..((.((((((((.....)))))))))).----------.((((((...(......)...)))))).....)).))))).......-. ( -28.30) >DroEre_CAF1 22782 113 + 1 ---AGUGGUUGUUUGGGCCGAAUGUUGGCUGAGUGAAGAGCCACGUGCAGUGCUUCUCAGUGGCUGCCA---GUGGCCAACGGGCCGCCAAGCCACUCAAACCAUCCAAACCACCCAAA ---.(((((((..((((((((...)))))((((((...((((((.((.((.....))))))))))((..---((((((....))))))...))))))))..)))..).))))))..... ( -47.60) >consensus ___AGUAG________________UUGGCUGAAUGAAGAGCCACGUGCAGUGGUUCUCAC__________UGGUGGCCAACGAGCCAGCAAGCCACCCAAAACACCCAAUCCAUCCA_A .........................(((((...((.((((((((.....))))))))))............(.((((......)))).).)))))........................ (-22.73 = -23.22 + 0.50)

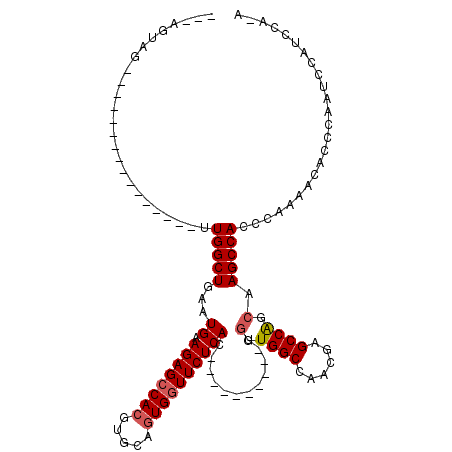
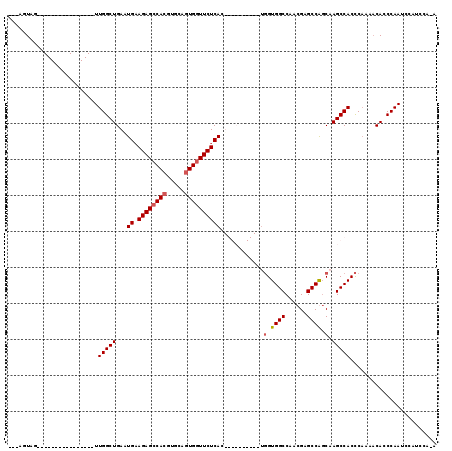
| Location | 1,572,005 – 1,572,107 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -23.20 |
| Energy contribution | -25.20 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1572005 102 - 23771897 U-UGGAUGGAUUGGGUGUUUGGGGUGGCUAGCUGGCUCGUUGGCCACCACUGGCAGCCACUGAGAACCACUGCACGUGGCUCUUCAUUCAGCCAA----------------CUACUUAC (-(((.((((((((.((((...((((((((((......))))))))))...)))).)))..((((.((((.....)))).)))).))))).))))----------------........ ( -41.70) >DroSec_CAF1 16484 82 - 1 --------GAUUGGGUGUUUUGGGUGGCUUGCCGGCUCGUUGGCCACCA----------GUGAGAACCACUGCACUUGGCUCUUCAUUCAGCCAA----------------CUACU--- --------...(((((((....(((((((.((......)).)))))))(----------(((.....))))))))((((((........))))))----------------)))..--- ( -27.30) >DroSim_CAF1 16667 89 - 1 U-UGGAUGGAUUGGGUGUUUUGGGUGGCUUGCUGGCUCGUUGGCCACCA----------GUGAGAACCACUGCACGUGGCUCUACAUUCAGCCAA----------------CUACU--- .-(((.(((.((((((((....(((((((.((......)).))))))).----------...(((.((((.....)))).)))))))))))))).----------------)))..--- ( -34.20) >DroEre_CAF1 22782 113 - 1 UUUGGGUGGUUUGGAUGGUUUGAGUGGCUUGGCGGCCCGUUGGCCAC---UGGCAGCCACUGAGAAGCACUGCACGUGGCUCUUCACUCAGCCAACAUUCGGCCCAAACAACCACU--- (((((((.(..((..(((((.((((((((.((.((((....)))).)---))))((((((((((.....)).)).))))))...)))))))))).))..).)))))))........--- ( -47.10) >consensus U_UGGAUGGAUUGGGUGUUUUGGGUGGCUUGCCGGCUCGUUGGCCACCA__________CUGAGAACCACUGCACGUGGCUCUUCAUUCAGCCAA________________CUACU___ ......(((.(((((((.....(((((((.((......)).))))))).............((((.((((.....)))).))))))))))))))......................... (-23.20 = -25.20 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:52 2006