
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,837,307 – 14,837,412 |
| Length | 105 |
| Max. P | 0.676469 |

| Location | 14,837,307 – 14,837,412 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -12.48 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14837307 105 + 23771897 UACAAAAAUAGAUGAAGAAAAAU---CCCCAAAAA-----AAGAGUAACCAAAACCAGACAACGGACCAAACUUGGCUCUGGCUUUGCCAUUGGCACUGUUGUAAUUUUCUAU ...............((((((..---.........-----..(.((.......)))..(((((((.((((...((((.........))))))))..)))))))..)))))).. ( -17.80) >DroPse_CAF1 70268 89 + 1 ---GAAACUACAAAAAACAAAAA----CGAG--UA-----AAGAGUAACCAA-ACCAAACAACGGACCAAAGUUGGCUCGAGCUUUGCCAUUGGCAUUGUUC---------AU ---....................----.(((--(.-----..(.((......-)))...((((........))))))))((((..((((...))))..))))---------.. ( -17.80) >DroSec_CAF1 14031 103 + 1 UACAAAAAUAGAUGAAGAAAAAU---CCCCA--AA-----AAGAGUAACCAAAACCAGACAACGGACCAAAGUUGGCUCUGGCUUUGCCAUUGGCAUUGUUGUAAUUUUCUAU ...............((((((..---.....--..-----..............(((((((((........))))..)))))(..((((...))))..)......)))))).. ( -16.80) >DroSim_CAF1 14586 103 + 1 UACAAAAAUAGAUGAAGAAAAAU---CCCAA--AA-----AAGAGUAACCAAAACCAGACAACGGACCAAAGUUGGCUCUGGCUUUGCCAUUGGCACUGUUGUAAUUUUCUAU ...............((((((.(---(....--..-----..))..............(((((((.((((...((((.........))))))))..)))))))..)))))).. ( -18.00) >DroYak_CAF1 10197 109 + 1 UACAAAAAUAGAUGAAGAAAACAAAGCC--A--AAAAAAGAAGAGUAACCAAAACCAGACAACGGACCAAAGUUGGCUCUGGCUUUGCCAUUGGCACUGUUGUAAUUUUCGAU ............((((((.((((..(((--(--(..((((.(((((...(.......).((((........)))))))))..))))....)))))..))))....)))))).. ( -21.70) >DroPer_CAF1 67715 89 + 1 ---GAAACUACAAAAAACAAAAA----CGAG--UA-----AAGAGUAACCAA-ACCAAACAACGGACCAAAGUUGGCUCGAGCUUUGCCAUUGGCAUUGUUC---------AU ---....................----.(((--(.-----..(.((......-)))...((((........))))))))((((..((((...))))..))))---------.. ( -17.80) >consensus UACAAAAAUAGAUGAAGAAAAAA___CCCAA__AA_____AAGAGUAACCAAAACCAGACAACGGACCAAAGUUGGCUCUGGCUUUGCCAUUGGCACUGUUGUAAUUUUCUAU ..........................................................(((((((.((((...((((.........))))))))..))))))).......... (-12.48 = -12.90 + 0.42)

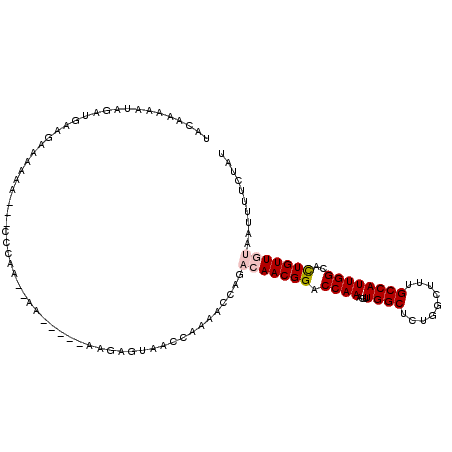
| Location | 14,837,307 – 14,837,412 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -16.81 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14837307 105 - 23771897 AUAGAAAAUUACAACAGUGCCAAUGGCAAAGCCAGAGCCAAGUUUGGUCCGUUGUCUGGUUUUGGUUACUCUU-----UUUUUGGGG---AUUUUUCUUCAUCUAUUUUUGUA (((((............((((...)))).(((((((((((..(..........)..)))))))))))......-----....(((((---(....)))))))))))....... ( -23.60) >DroPse_CAF1 70268 89 - 1 AU---------GAACAAUGCCAAUGGCAAAGCUCGAGCCAACUUUGGUCCGUUGUUUGGU-UUGGUUACUCUU-----UA--CUCG----UUUUUGUUUUUUGUAGUUUC--- ..---------((((((((((...)))).(((.(((((((((..((...))..).)))))-))))))......-----..--....----...))))))...........--- ( -15.80) >DroSec_CAF1 14031 103 - 1 AUAGAAAAUUACAACAAUGCCAAUGGCAAAGCCAGAGCCAACUUUGGUCCGUUGUCUGGUUUUGGUUACUCUU-----UU--UGGGG---AUUUUUCUUCAUCUAUUUUUGUA (((((.........(((.((((..(((((.(((((((....)))))))...))))))))).))).........-----..--(((((---(....)))))))))))....... ( -26.00) >DroSim_CAF1 14586 103 - 1 AUAGAAAAUUACAACAGUGCCAAUGGCAAAGCCAGAGCCAACUUUGGUCCGUUGUCUGGUUUUGGUUACUCUU-----UU--UUGGG---AUUUUUCUUCAUCUAUUUUUGUA ..((((((......(((.((((..(((((.(((((((....)))))))...))))))))).)))....(((..-----..--..)))---..))))))............... ( -23.30) >DroYak_CAF1 10197 109 - 1 AUCGAAAAUUACAACAGUGCCAAUGGCAAAGCCAGAGCCAACUUUGGUCCGUUGUCUGGUUUUGGUUACUCUUCUUUUUU--U--GGCUUUGUUUUCUUCAUCUAUUUUUGUA ..(((((((...(((((.(((((.(.((((((((((..((((........)))))))))))))).).............)--)--))).)))))..........))))))).. ( -24.86) >DroPer_CAF1 67715 89 - 1 AU---------GAACAAUGCCAAUGGCAAAGCUCGAGCCAACUUUGGUCCGUUGUUUGGU-UUGGUUACUCUU-----UA--CUCG----UUUUUGUUUUUUGUAGUUUC--- ..---------((((((((((...)))).(((.(((((((((..((...))..).)))))-))))))......-----..--....----...))))))...........--- ( -15.80) >consensus AUAGAAAAUUACAACAAUGCCAAUGGCAAAGCCAGAGCCAACUUUGGUCCGUUGUCUGGUUUUGGUUACUCUU_____UU__UUGGG___AUUUUUCUUCAUCUAUUUUUGUA ..............(((.((((..(((((.(((((((....)))))))...))))))))).)))................................................. (-16.81 = -16.62 + -0.19)

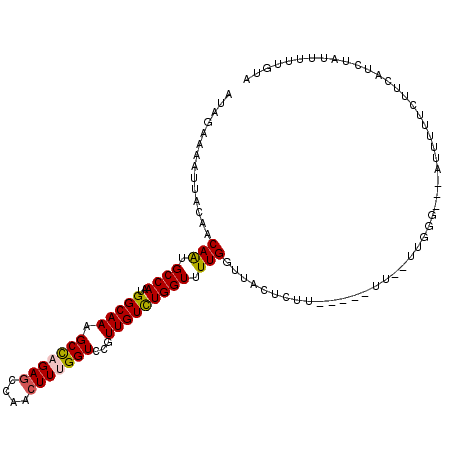
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:51 2006